Method for detecting CVM deleterious gene of oxen
A technology of harmful genes and genotypes, which is applied in the fields of biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of unclear carrying status of harmful genes in CVM, and achieve the effect of simple method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Design of primers and optimization of amplification conditions
[0022] According to the bovine SLC35A3 gene sequence (NO: AY160683) on GenBank, Oligo6.0 was used to design specific PCR primers for detecting CVM harmful genes. , screening and optimization, and finally determine the following primer sequences:
[0023] CVMF (upstream): 5-TGGCCCTCAGATTCTCAAGAG-3'
[0024] CVMR (downstream): 5-CCAAGTTGAATGTTTCTTATC-3'
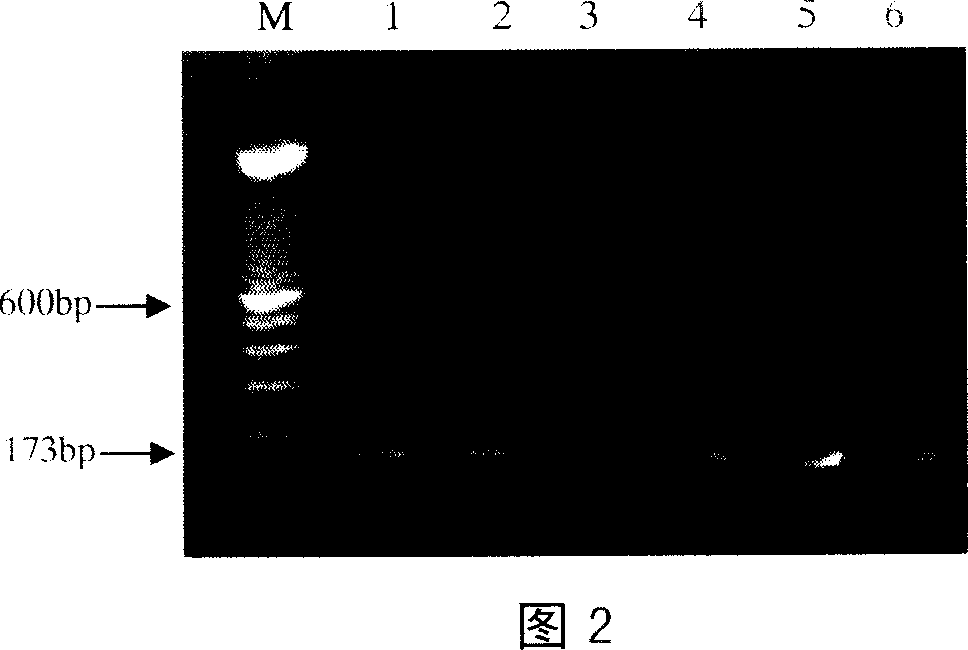
[0025] The optimal conditions for PCR primers were: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 55±10°C for 30 s, extension at 72°C for 30 s, and 35 cycles; extension at 72°C for 7 min. The detection result of agarose electrophoresis with optimized PCR conditions is shown in Figure 1. According to the detection results with optimized PCR conditions, the annealing temperature of the primer was determined to be 57°C.
[0026] The reaction system is: 25 pmol of primers, 50 ng of template DNA, 0.5 U of Taq enzyme...
Embodiment 2
[0028] Example 2 Preparation and electrophoresis of 12% non-denaturing polyacrylamide gel
[0029] 1. Clean the glass plate, dry it, and seal the gap between the glass plate and the strip with 0.8% agarose;
[0030] 2. Prepare 30ml of 12% acrylamide glue working solution (12.0ml of 30% acrylamide, 3ml of 50% glycerin, 3ml of 10×TBE, 200μl of 10% ammonium persulfate, 12μl of TEMED, 12ml of pure water), and pour the glue quickly after mixing;
[0031] 3. Stop pouring glue when pouring is 0.1cm away from the edge of the glass plate, insert a comb, polymerize at room temperature for about half an hour, and store excess acrylamide at 4°C. And observe the gel polymerization at any time, add acrylamide;
[0032] 4. After the gel is polymerized, add 1×TBE to the electrophoresis tank, and rinse the sample hole with a syringe;
[0033] 5. Pre-electrophoresis for 20 minutes, and prepare for loading at the same time;
[0034] 6. Take 1.5 μl of PCR product and place it in a PCR tube, ad...
Embodiment 3
[0036] The PCR-SSCP analysis of embodiment 3 CVM
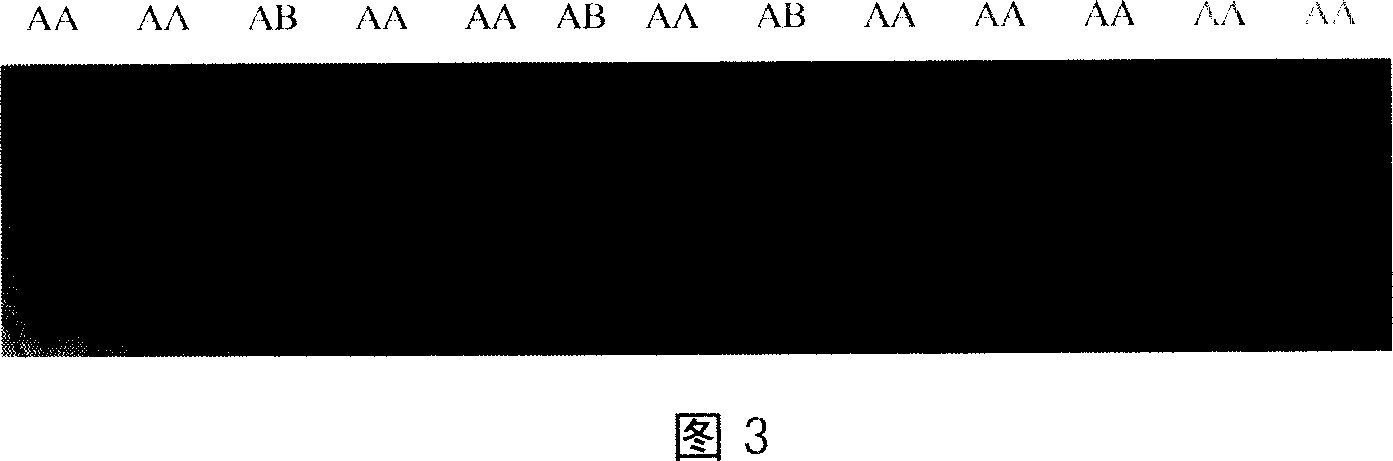
[0037] Utilize the DNA sample of bull, the selected specificity PCR primer CVMF / CVMR in embodiment 1 to carry out PCR amplification, this amplified fragment comprises the G→T base mutation that causes CVM, gets 1.5 μ l PCR products and places PCR tube Add 6 μl of denaturing Buffer, denature at 98°C for 10 min, place it on ice immediately until electrophoresis, use the method in Example 2 to make a 12% polyacrylamide gel with acr:bis of 29:1 for electrophoresis, 120V, 14 -16h, after gel electrophoresis, use the silver staining method of Bassam et al. (1991) to stain [9] , found that there are two different genotypes: AA type and AB type, the genotyping results are shown in Figure 3. After sequencing, it was found that the 73rd base of the 4th exon (exon4) of the AA-type SLC35A3 gene (sequence identical to SEQ ID NO.3) was G, and this genotype belonged to a normal individual, represented by "TV"; and The 73rd base of exon 4 (e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com