PCR detection method for vibrio cholerae, vibrio parahaemolyticus, and vibrio mimicus in food
A technology for Vibrio cholerae and Vibrio hemolyticus, which is applied in biochemical equipment and methods, microbial determination/inspection, measurement devices, etc., can solve problems such as never seen before, and achieves reduction of detection cost, acceleration of detection speed, and reduction of The effect of workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment one: the detection of cuttlefish
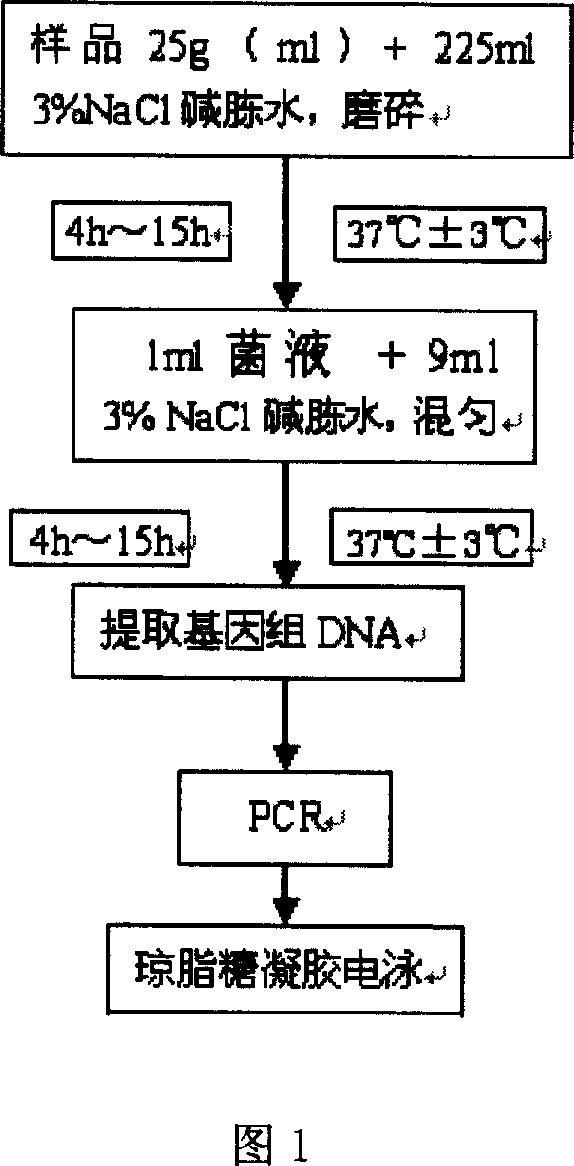
[0024] 1. Aseptic operation, take 25g breaded shrimp sample, cut it into pieces, put it into a sterilized container filled with 225ml 3% NaCl alkaline peptone water (APW), and incubate at 37°C±1°C for 4h-15h.
[0025] 2. Take 1ml of the enrichment solution and add it to a sterilized test tube filled with 9ml of 3% NaCl alkaline peptone water (APW), and incubate at 37°C±1°C for 4h to 15h.
[0026] 3. Pipette gun to absorb 1.5ml of the cultured bacterial solution, add it to a 1.5ml centrifuge tube and centrifuge at 13000rpm for 1min, discard the supernatant.
[0027] 4. Add 0.6ml of sterile deionized water to suspend the precipitate, shake and mix well, and bathe in 100°C water bath for 10min.
[0028] 5. Centrifuge at 12000 rpm for 5 minutes, take the supernatant as a DNA template, and store it at -20°C for later use.
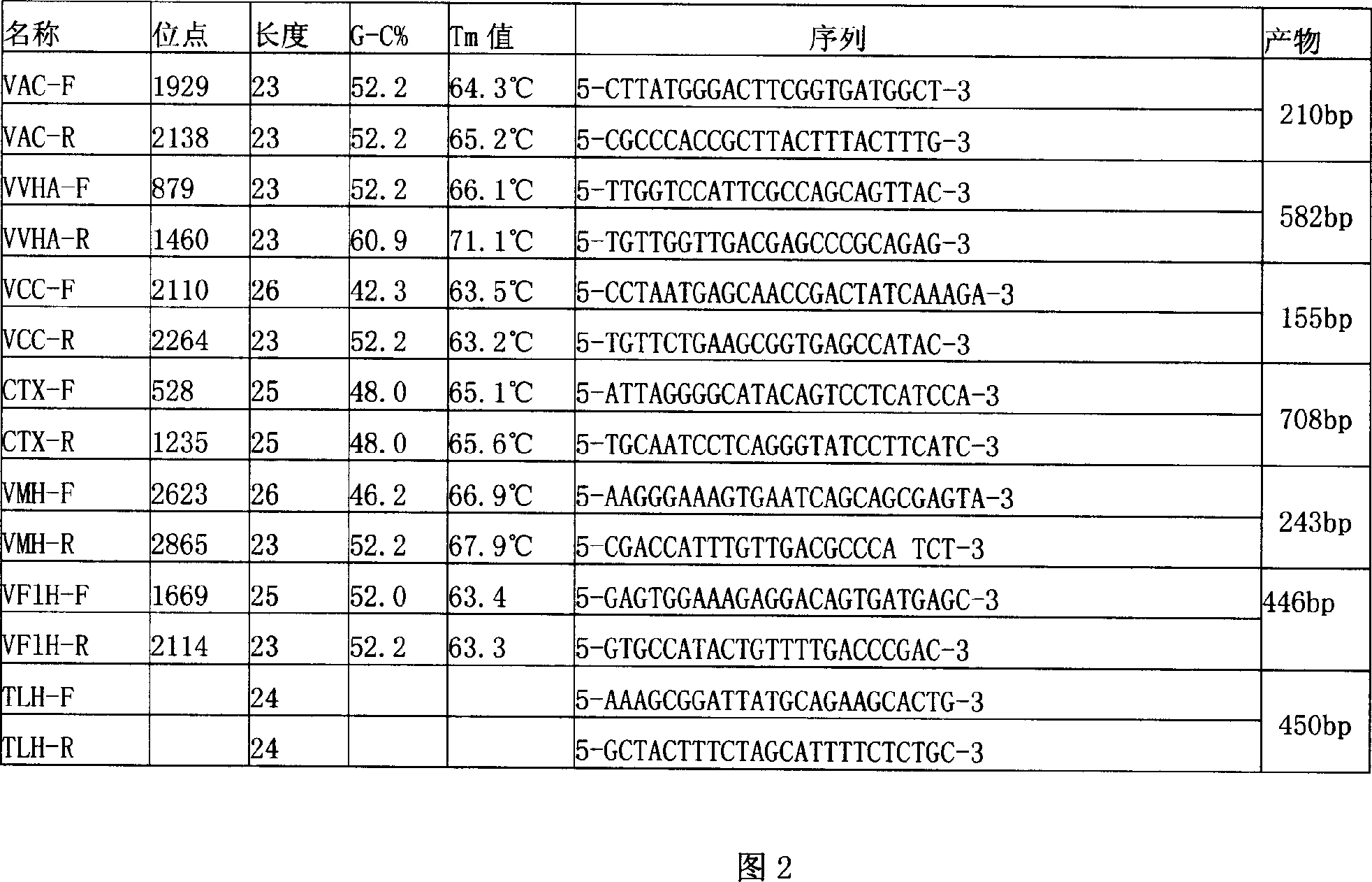
[0029] 6. Add sample and do PCR
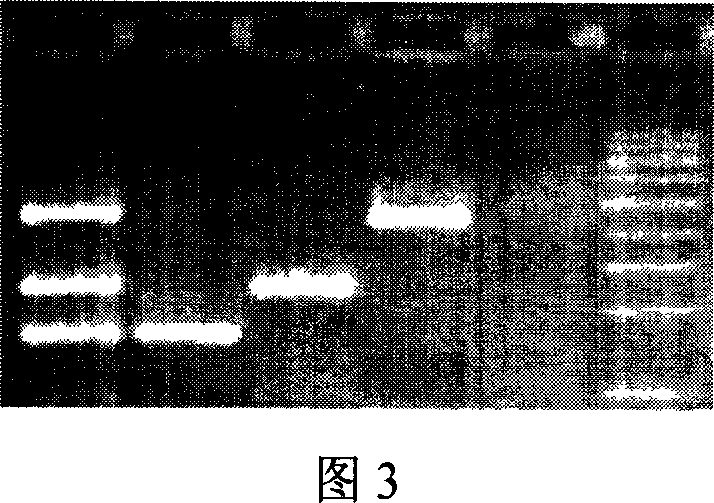
[0030] 7. Take 10 μl of the PCR reaction solution for agarose gel elec...
Embodiment 2
[0033] Embodiment two: the detection of soft-shelled turtle eggs
[0034] 1. Aseptic operation, take 25g clam sample, cut it into pieces, put it into a sterilized container filled with 225ml 3% NaCl alkaline peptone water (APW), and incubate at 37°C±1°C for 4h-15h.
[0035] 2. Take 1ml of the enrichment solution and add it to a sterilized test tube filled with 9ml of 3% NaCl alkaline peptone water (APW), and incubate at 37°C±1°C for 4h to 15h.
[0036] 3. Pipette gun to absorb 1.5ml of the cultured bacterial solution, add it to a 1.5ml centrifuge tube and centrifuge at 13000rpm for 1min, discard the supernatant.
[0037] 4. Add 0.6ml of sterile deionized water to suspend the precipitate, shake and mix well, and bathe in 100°C water bath for 10min.
[0038] 5. Centrifuge at 12000 rpm for 5 minutes, take the supernatant as a DNA template, and store it at -20°C for later use.
[0039] 6. Add sample and do PCR
[0040] 7. Take 10 μl of the PCR reaction solution for agarose gel ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com