Promoters and proteins from Clostridium thermocellum and uses thereof
a technology of clostridium thermocellum and promoters, applied in the field of nucleic acid promoters and proteins, can solve the problems of limiting conversion yield, complicated task of elucidating the regulatory mechanism, and long list of cellulosomal genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning of glyR3 Nucleic Acid Molecule
[0105] A set of primers were synthesized (Invitrogen, Carlsbad, Calif.) to amplify the glyR3 gene, as well as, to add EcoRV and XhoI restriction sites for cloning: (SEQ ID NO:8) glyR3-F-EcoRV-GCGCGATATCACCAGTGAAGAAATAGCAAAATTA; (SEQ ID NO:9) glyR3-R-XhoI-GCGCCTCGAGGAATTCCAAAGCCCTCTTGGTT
[0106] Polymerase chain reaction (PCR)was utilized using genomic C. thermocellum DNA as a template. Extensor Hi-Fidelity PCR Enzyme (ABgene) was the polymerase of choice due to its ability to accurately amplify longer DNA products. The standard Extensor protocol was followed with the exception of an 80 second PCR extension time. The PCR product was run on a 1% agarose gel with a molecular weight marker to verify the correct size. Next, the PCR product was digested with EcoRV and XhoI. The PTXB 1 plasmid was digested with NruI and XhoI. The products of the digestions were ligated. The ligation product was transformed by electroporation (Bio-Rad, Hercules, Calif. ...
example 2
Expression and Purification of Recombinant Protein
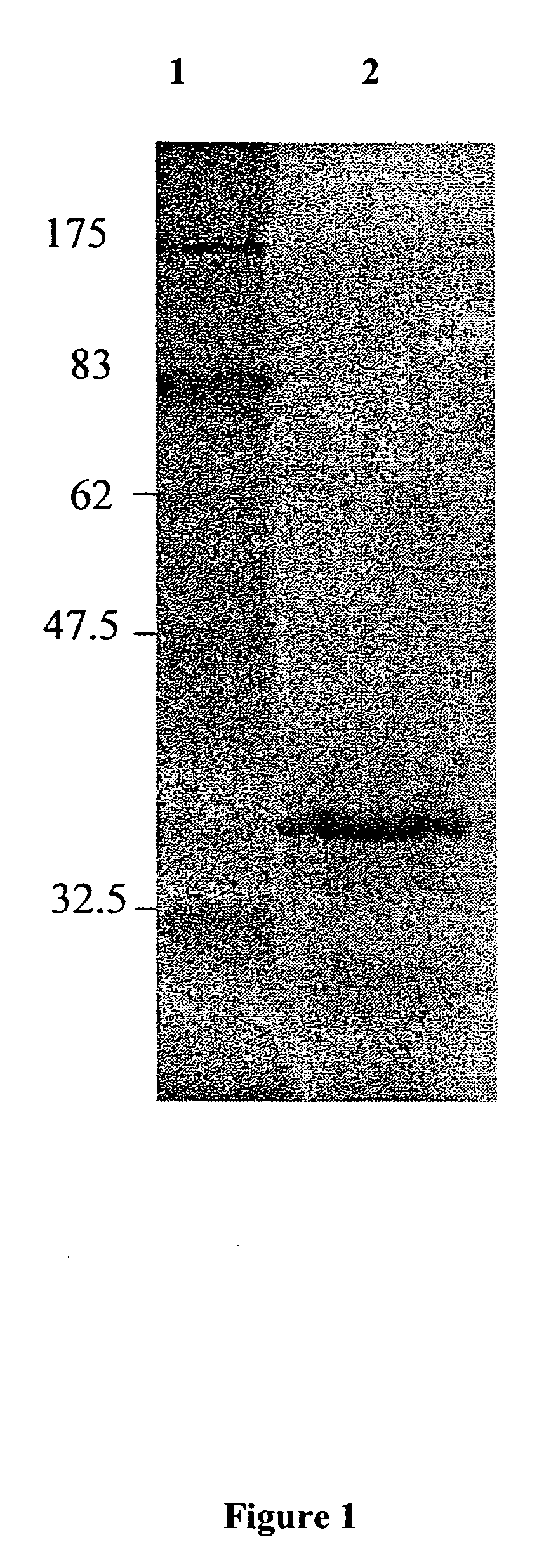
[0107] PTXB 1 containing the glyR3 insert was transformed by electroporation into E. Coli BL21DE3. cells. The cells were grown to a density of 0.8 (OD 600), then 50 mM IPTG was added to the culture. After inducing the expression of rGlyR3 with IPTG, the culture was allowed to incubate in a shaker at 37° C. for 4 hours. At 4 hours, the culture was centrifuged at 5,000 g for 5 minutes and the supernatant decanted. The New England Biolabs (NEB) IMPACT system protein purification protocol was followed. The cells were resuspended in column buffer (20 mM HEPES, 500 mM NaCl, and 1 mM EDTA) and then sonicated for cell lysis. The sonicated product was centrifuged and the supernatant was added to chitin beads (NEB) at room temperature for 1 hour. The chitin beads were washed with 200 ml of column buffer at a flow rate of 2 ml / min. Next, the beads were incubated with 100 mM DTT at 4° C. overnight. The resulting flow-through was concentrated us...
example 3
Creating DNA Probes for EMSA
[0108] Probes for EMSA were created using PCR with Thermostart Taq (ABgene) as a polymerase. The standard Thermostart protocol was used with varying extension times (1 kb / minute rule always followed) and different annealing temperatures (57° C.-62° C.). Primers were synthesized with a 5′ biotin label by Invitrogen (Carlsbad, Calif.). Primers (5′-3′) used:
(SEQ ID NO:10)entire_celCProm-F-biotin:- CCGAATAAAAACTGGACAGAG;(SEQ ID NO:11)Entire_celCProm-R-unlab:- TCCTCCTGAAATATTGTGTTTTA(SEQ ID NO:12)celCProm_1st_100bp-R-unlab:-TGAAACCATTTAACACTGGATTAT(SEQ ID NO:13)celCProm_2nd_100bp-F_biotin-GTTTACGATTTCAAATGTTTATATC.
[0109] For probes that contained just the 18 bp binding site, complementary DNA fragments were synthesized and annealed by heating to 94 C and then cooling:
(SEQ ID NO:14) BS-F-Biotin- AATGAACGCGCGTACATT(SEQ ID NO:15) BS-R-Unlab- AATGTACGCGCGTTCATT
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| extension time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com