Lectin-derived progenitor cell preservation factors and methods of use
a technology of progenitor cells and preservation factors, applied in the field of nuclear acid, can solve the problems of inability to easily be clinically applied, no known cytokines alone or in combination can preserve the viability of primitive progenitors in liquid culture without stromal support, and the efficiency of transfection is not easily determined. , to achieve the effect of increasing the level of total body irradiation, increasing the targeting efficiency, and determining the efficiency of transfection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
RNA Isolation and cDNA Synthesis
[0132] Total RNA was prepared from mid-maturation Dolichos lab lab seeds stored at −70° C. following the procedure of Pawloski et al. (1994). Poly(A+) RNA was obtained from this total RNA using the PolyATract mRNA Isolation System (Promega) according to the manufacturer's instructions. Avian myeloblastosis virus reverse transcriptase (Promega) was used to generate cDNA from 0.5 μg poly(A+)RNA, or from 3.0 μg of total RNA, using 1 μg of oligo(dT) in standard reaction conditions (Sambrook et al. 1989).
example 2a
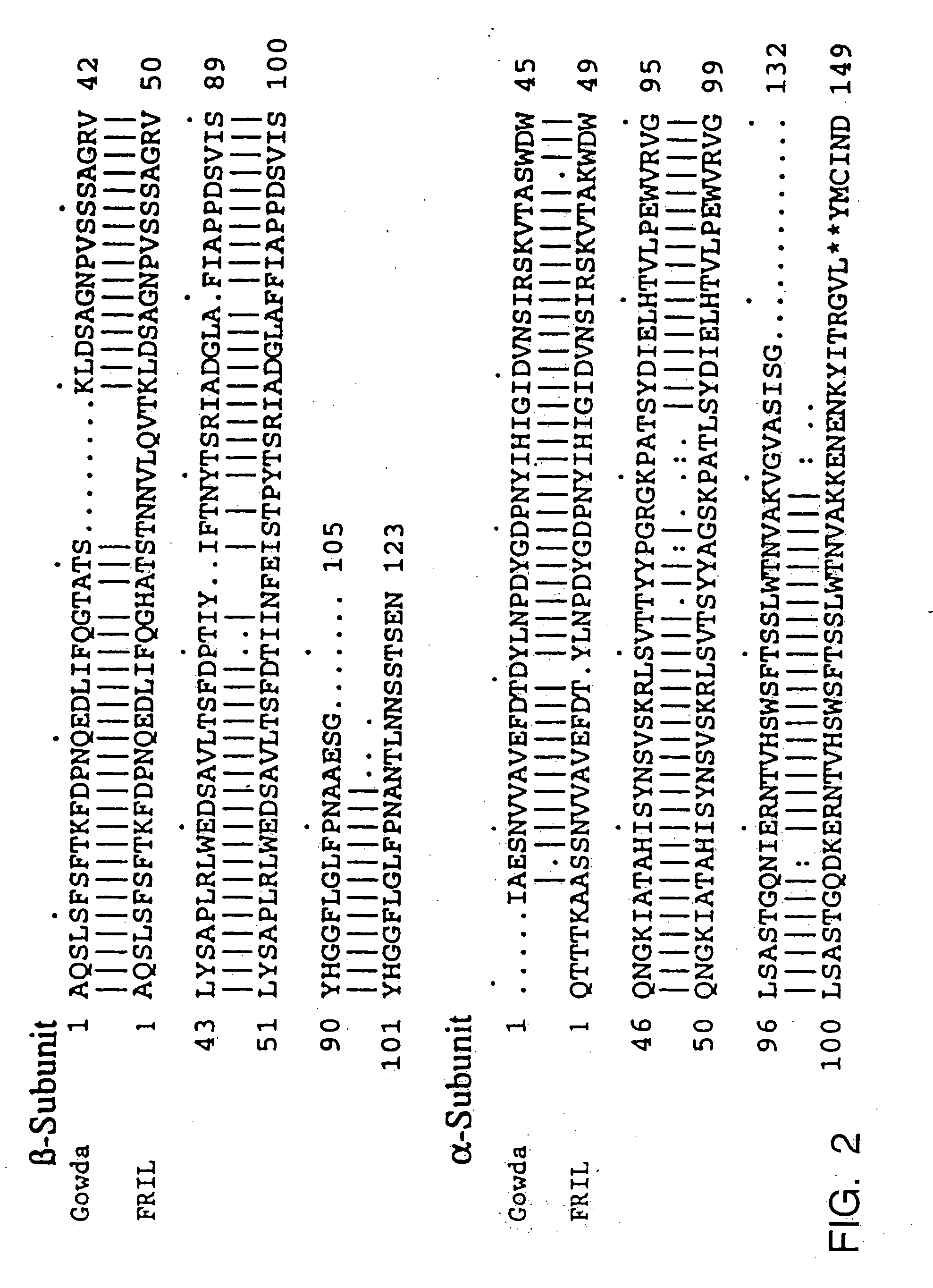
[0133] Based on the amino acid sequence published by Gowda et al. (1994), two degenerate oligonucleotide primers were designed using Phaseolus codon usage (Devereux et al. 1984):
MLAAA (AG) TT (TC) GA (TC) CC (AT) AA(SEQ ID NO:3)(TC) CA (AG) GA (AG) GAMLZTT (AT) CC (AG) TT (TC) TGCCA (AG)(SEQ ID NO:4)TCCCA
[0134] A 500+ bp product was amplified from cDNA prepared as described in Example 1, by 30 cycles of polymerase chain reaction (PCR), each cycle comprising 40 s at 94° C., 40 s at 50° C., 60 s at 72° C., followed by an extension step at 72° C. for 10 min. Reactions were performed in 50 μL containing 30 pmol of each primer, 0.2 mM deoxyribonucleotides and 0.5 unit of AmpliTaq polymerase (Perkin Elmer) in the corresponding buffer.
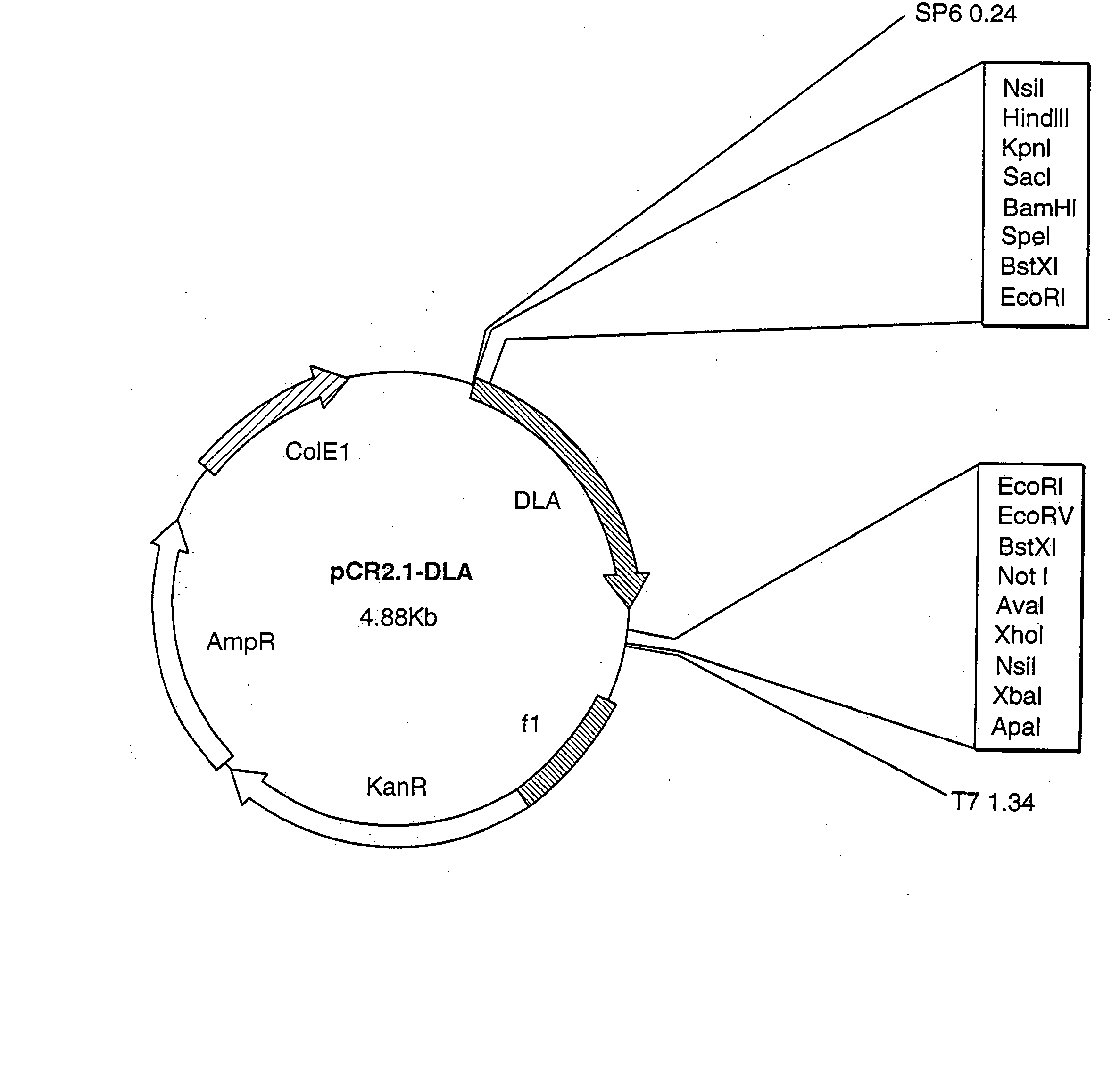
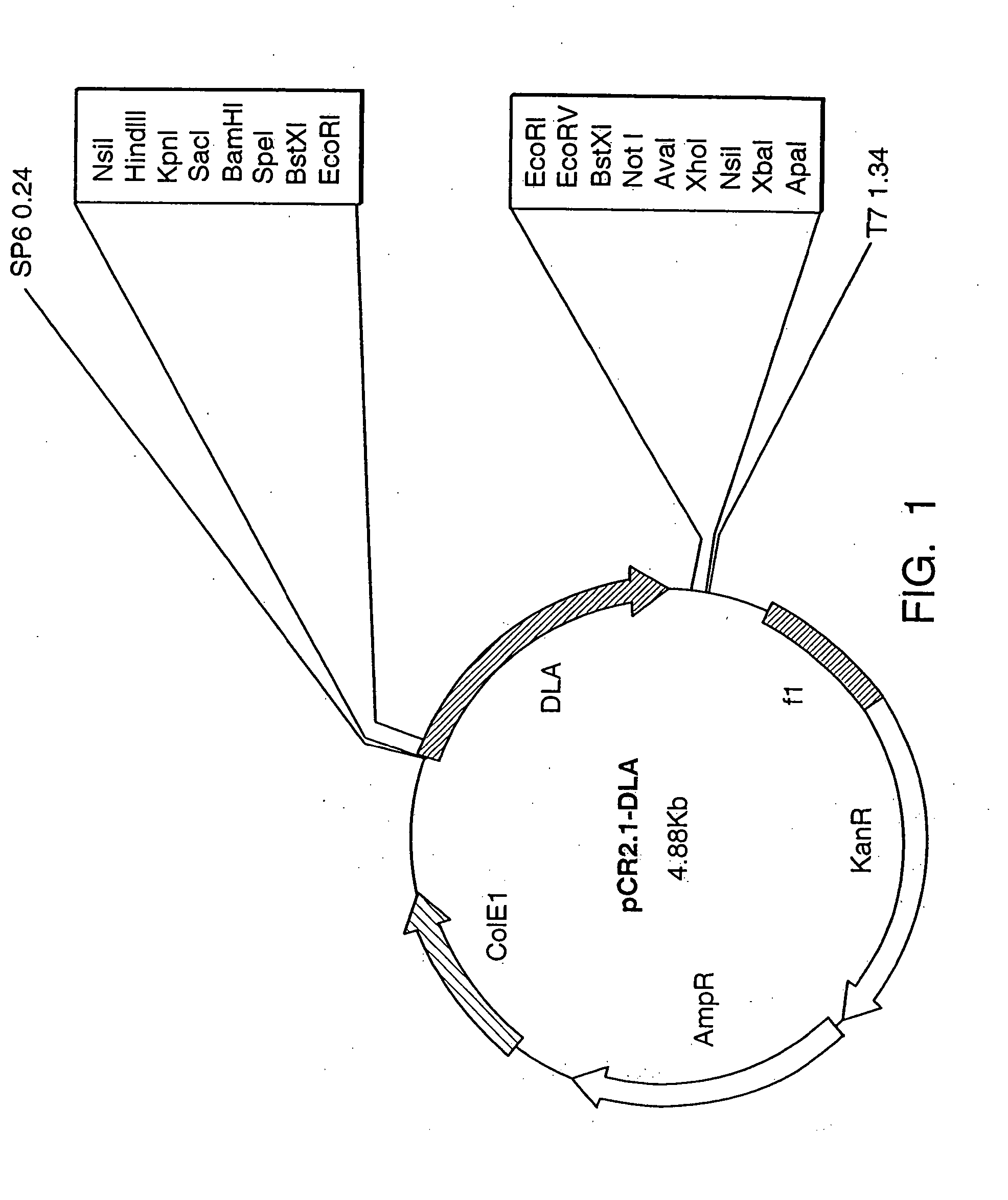
[0135] The 500 bp product obtained by PCR was cloned in the cloning vector, pCR2.1 (Invitrogen), and sequenced by sequenase dideoxy chain termination (United States Biochemicals) using the following primers:
GTACCGAGCTCGGAT(SEQ ID NO:5)TCTAGATGCATGCTCGAG(...
example 2b
[0136] Based on the sequence of the FRILa amplified product, a specific primer was prepared:
MLXGTTGGACGTCAATTCCGATGTG(SEQ ID NO:7)
[0137] A degenerate primer corresponding to the first five amino acids of the sequence published by Gowda et al. (1994) was also prepared:
MLIGC (TC) CA (AG) TC (TC) CT (TC) TC(SEQ ID NO:8)(TC) TT
[0138] The MLX and MLI primers were used in combination to amplify a 480 bp product from cDNA prepared as in Example 1, through 30 PCR cycles using the same conditions described above. This secondary amplified fragment was cloned in the pCR2.1 vector and sequenced as described above, and was designated “FRILb.”
PUM
| Property | Measurement | Unit |
|---|---|---|
| Therapeutic | aaaaa | aaaaa |
| Cytotoxicity | aaaaa | aaaaa |
| Affinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com