Method for producing, in yeast, a hydroxylated triple helical protein, and yeast host cells useful in said method
a technology of which is applied in the field of producing, in yeast, a hydroxylated triple helical protein and yeast host cells useful in said method, can solve the problems of many episomal type vectors suffering stability problems, their use risks serious immunogenicity problems, and the transmission of infective diseases and spongiform encephalopathies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
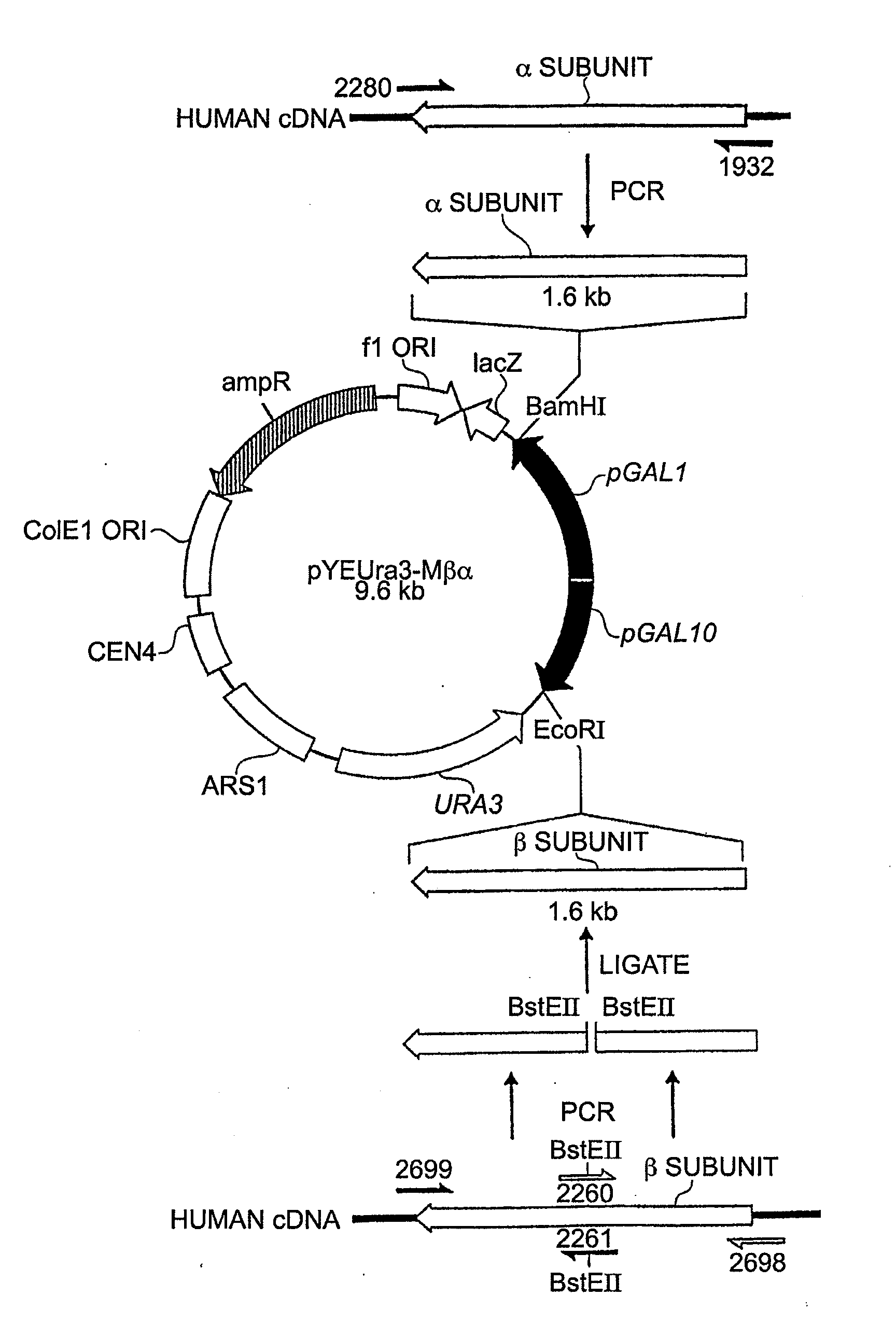
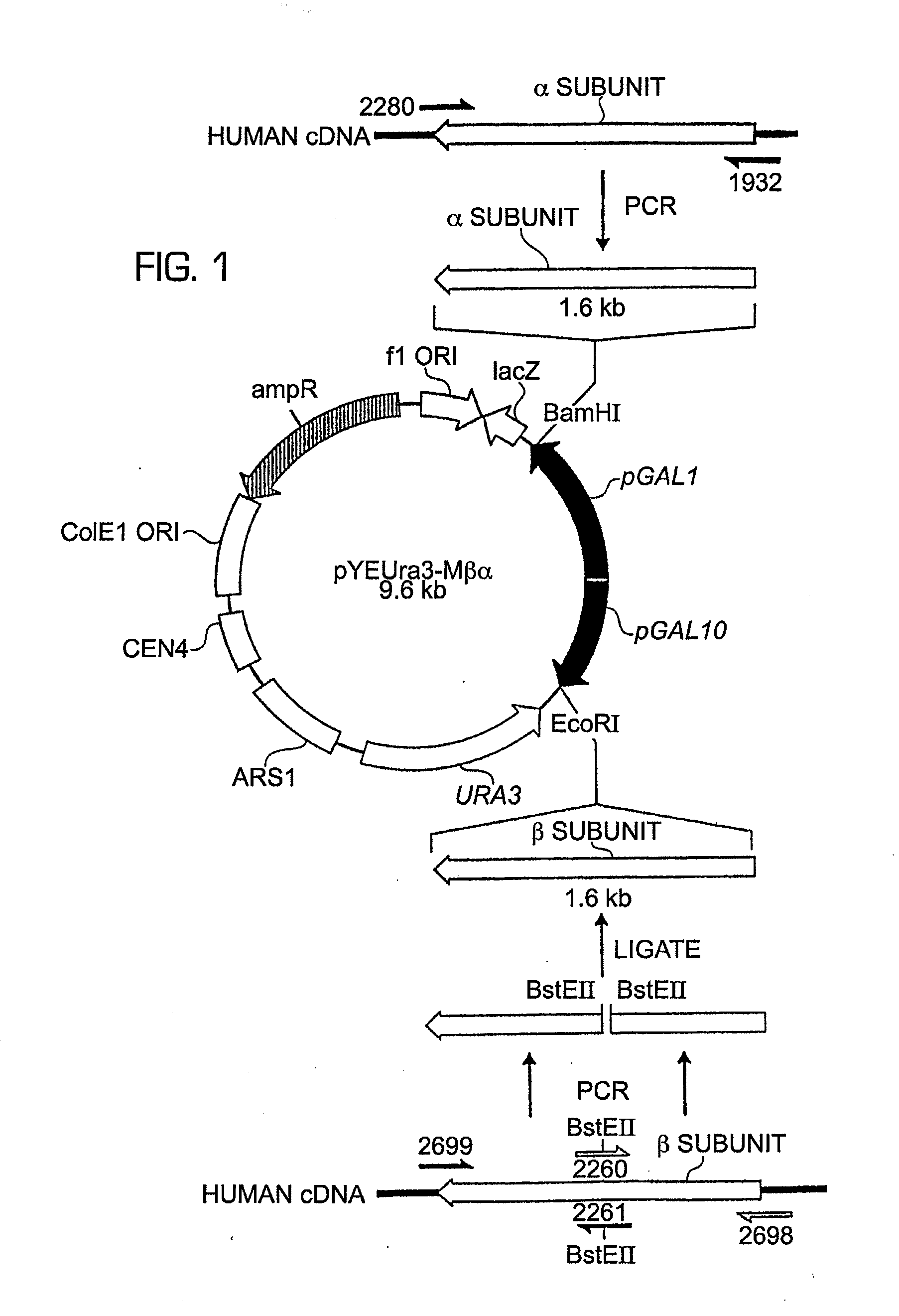
Construction of a Yeast Vector for Co-Ordinated Co-Expression of the a and β Subunits of Prolyl-4-Hydroxylase
Production of Yeast Expression Vector:
[0078]pYEUra3 (Clontech) contains the bidirectional promoter for GAL 1-10 expression. Induction by galactose in the absence of glucose results in high level expression from pGAL1 of any protein encoded by DNA sequences inserted in the correct orientation in the MCS (multiple cloning site) [either XhoI, SalI, XbaI or BamHI sites] provided there is an initiating ATG start codon. For pGAL10, expression induced by galactose occurs if the DNA sequences to be expressed are inserted in frame with the ATG codon of GAL 10 when said DNA sequences to be expressed is inserted in the EcoRI site.
[0079]In order to utilise the EcoRI site for cloning, without the necessity that the insert be in frame with the ATG of GAL10 for expression, it was necessary to modify pYEUra3 to remove the GAL10 initiation codon. This was done as follows. A PCR fragment was g...
example 2
Co-ordinated Co-Expression of a Collagen Segment and Prolyl-4-Hydroxylase (α and β Subunit) and Synthesis of Hydroxylated Collagen TYPE III in Yeast
[0083]A 1.6 kbp recombinant collagen fragment was generated by PCR using primers 1989 [Forward primer 5′-gct.agc.aag.ctt GGA.GCT.CCA. GGC.CCA.CTT.GGG.ATT.GCT.GGG-3′ (SEQ ID NO: 15)] and 1903 [Reverse primer 5′-tcg.cga.tct.aga.TTA.TAA.AAA.GCA.AAC.AGG.GCC.AAC.GTC.CAC. ACC-3′ (SEQ ID NO:16)] homologous to a region of the collagen type III alpha 1 chain (COL3A1). The template for isolation of the fragment of type III collagen alpha 1 chain was prepared from Wizard purified DNA obtained from a cDNA library [HL1123n Lambda Max 1 Clontech Lot#1245, Human Kidney cDNA 5′-Stretch Library].
[0084]The actual size of the isolated 1.6 kbp fragment is 1635 bp, comprising 1611 bp of COL3A1 DNA flanked either side by 12 bp derived from the primers. The 1611 bp of COL3A1 DNA corresponds to nucleotides #2713-4826 (i.e. codon #905-1442) of the full-length co...
example 3
Use of Yeast Artificial Chromosomes [YACs] for Co-Ordinated expression of the α and β Subunits of Prolyl-4-Hydroxylase [P4H]
[0101]pYAC5 [11454 bp] (Kuhn and Ludwig, 1994) was digested with BamHI to liberate the HIS3 gene [1210 bp] from between the 2 telomere ends and with SalI-NruI to produce two fragments [left arm: fragment 1, 5448 bp & right arm: fragment2, 4238 bp] which were gel purified. Fragment 1 was BamHI-telomere end-E. coli ori-β-lactamase gene [ampicillin-resistance]-TRP1-ARS1-CEN4-tRNAsup-o-SalI. Fragment 2 was BamHI-telomere end-URA3-NruI.
[0102]pYEUra3.2.12β#39α#5 was digested with SalI-EcoRV to produce a P4H expression cassette fragment of the form SalI-XbaI-BamHI-α-ATG-BamHI-pGAL1-10-EcoRI-ATG-β-EcoRI-SmaI-EcoRV [4864 bp] which was gel purified. The expression cassette fragment encoding the cc and 13 subunits of P4H under the control of a galactose inducible bidirectional promoter was ligated with fragments 1 and 2 of the BamHI-SalI-NruI digested pYAC5 and the ligati...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com