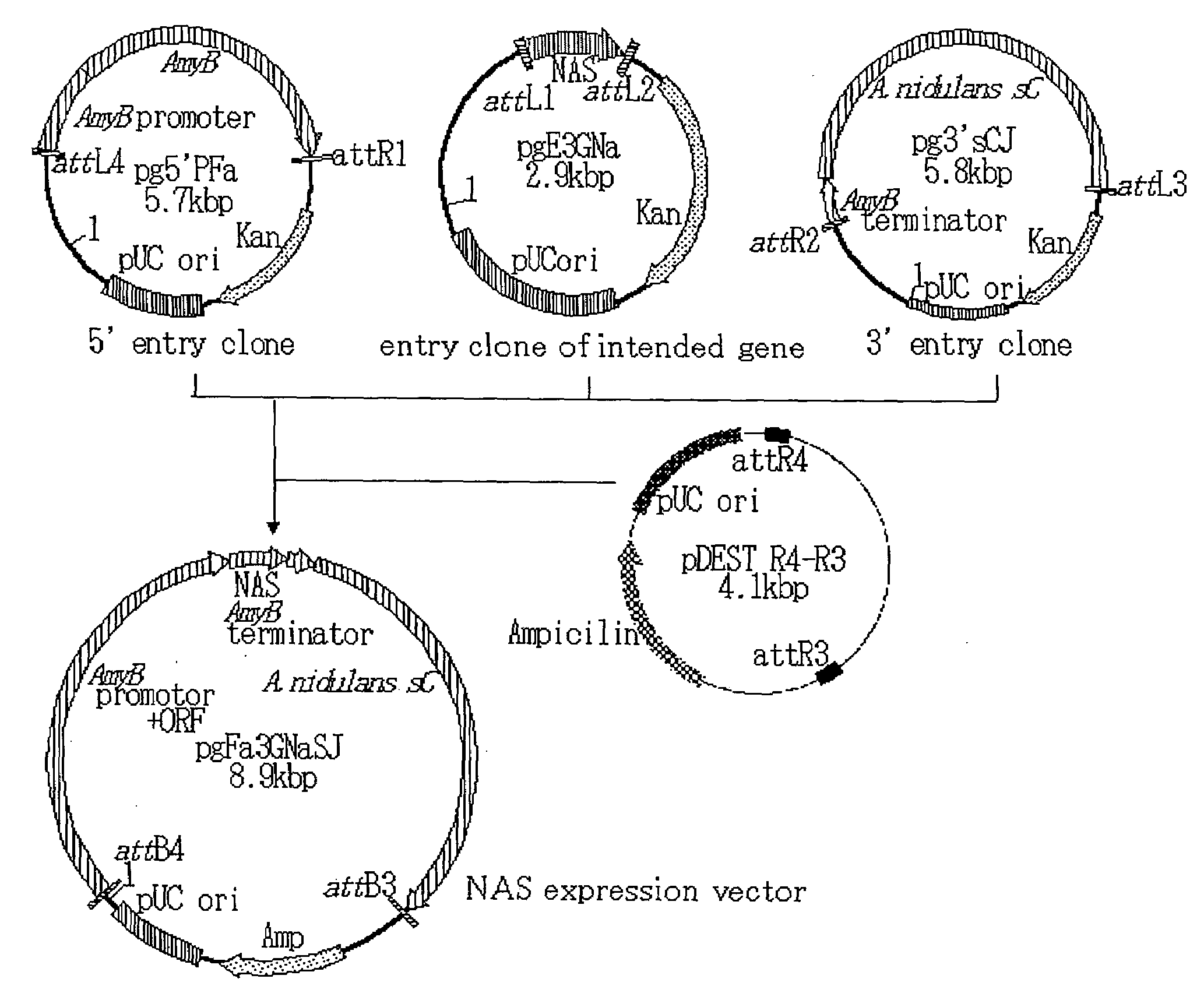
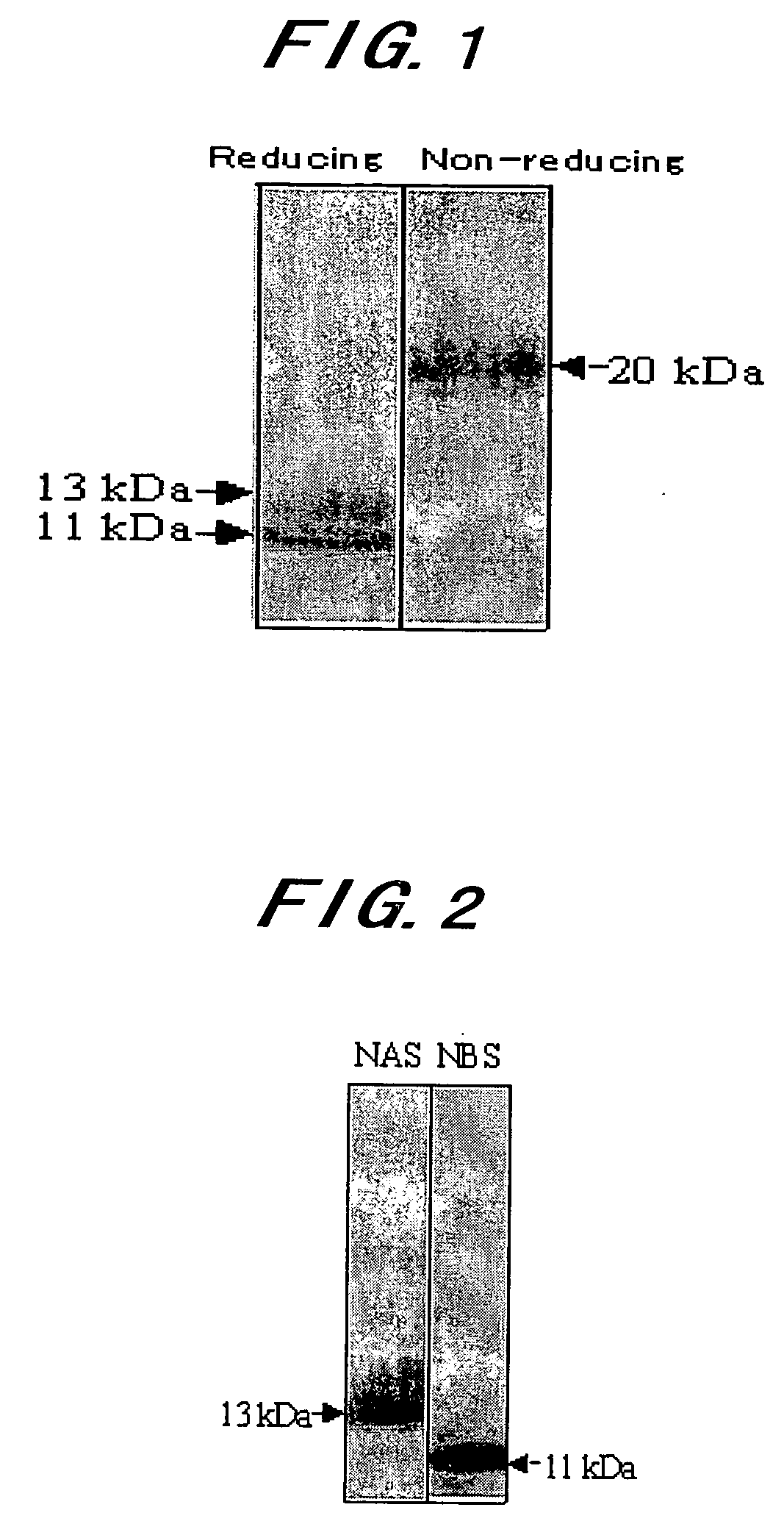
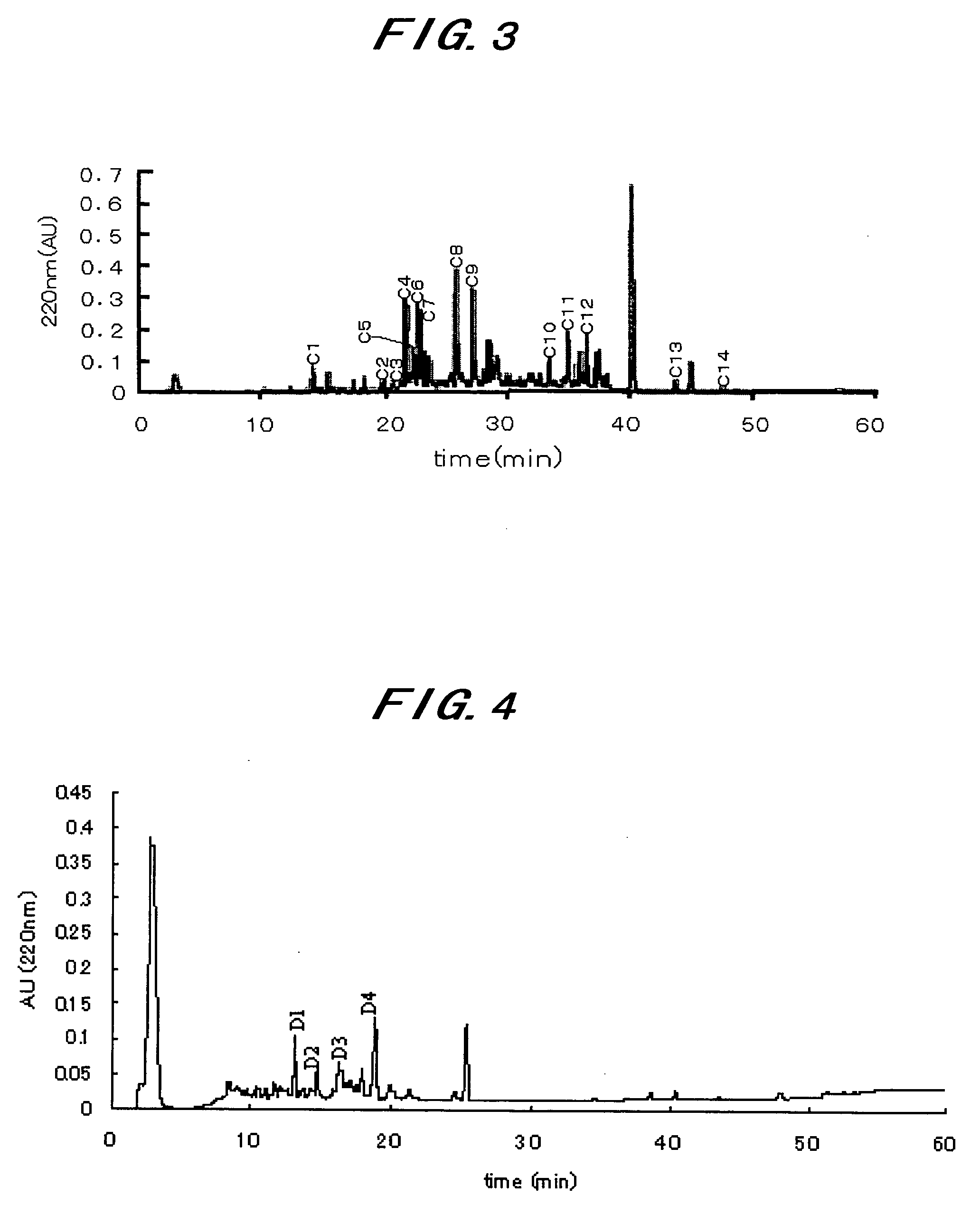
Novel Taste-Modifying Polypeptide Nas, Dna Thereof and Use Thereof
a technology of taste-modifying polypeptides and dna, which is applied in the direction of peptides, plant/algae/fungi/lichens ingredients, and peptides, etc., can solve the problems of insufficient taste-modifying functions of such reagents, disadvantages in laborious work and production costs, and inability to use such reagents in industrial production, etc., to achieve excellent taste-modifying activity and protein supply efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0123]A fruit of Curculigo latifolia was purified by the following procedures to obtain the novel protein with the taste-modifying activity.
(1) Preparation of Crude Extract Solution
[0124]40 liters of pure water were added to about 1 kg of the freeze-dried fruit of Curculigo latifolia (freeze-dried fruit powder in Table 1) for homogenization for 15 minutes. Then, centrifugation at 6,000 rpm for 20 minutes was done to discard the supernatant (the supernatant had no taste-modifying activity). The procedure described above was repeated twice, to obtain the residual precipitate.
[0125]Then, 20 liters of 0.05N sulfuric acid were added to the residual precipitate, for homogenization for 10 minutes. Then, centrifugation at 6,000 rpm for 20 minutes was done to recover the supernatant. The procedure described above was repeated twice. The resulting precipitate had no taste-modifying activity.
[0126]Then, 2 liters of 1N sodium hydroxide were added to the extract solution for neutralization, to o...
example 2
[0134]Neoculin obtained in Example 1 was purified further by the following procedures, to make the analysis of the individual subunits constituting neoculin.
(1) Purification on HiTrap SP Sepharose Fast Flow Column
[0135]100 mg of the neoculin powder obtained in Example 1 was dissolved in 20 ml of buffer A (50 mM Tris-HCl buffer, pH 7.5 containing 8M urea and 30 mM DTT). Then, the whole volume was applied to HiTrap SP Sepharose Fast Flow column (manufactured by Amersham Biosciences; a 1.6 cm diameter×2.5 cm) equilibrated with the buffer A. Continuously, the column was washed with 50 ml of the buffer A. Continuously, elution was done with 50 ml of the buffer A containing 1M NaCl, to obtain a purified NBS fraction. Furthermore, 70 ml of the wash fraction was dialyzed against ion exchange water to a volume of 100 ml.
(2) Second Purification on HiTrap SP Sepharose Fast Flow Column
[0136]To 100 mg of the wash fraction obtained above in (1) was added 8 M urea, 30 mM DTT and 50 mM acetate buff...
example 3
[0139]The amino acid sequence of the NAS fraction constituting neoculin as obtained in Example 2 was analyzed by the following procedures.
(1) Analysis of N-Terminal Amino Acid Sequence
[0140]70 μg of the purified NAS powder obtained in Example 2(1) to (3) was subjected to two-dimensional electrophoresis. The gel after electrophoresis was overlaid on a polyvinylidene difluoride (PVDF) membrane, where an electric current passed vertically for transfer. The PVDF membrane after the transfer was stained with SYPRO Ruby protein blot stain (manufactured by Molecular Probes), from which bands were excised out by a general method for the analysis of the N-terminal amino acid sequence with an amino acid sequencer (HP G1005A Protein Sequencing System). In other words, phenylisothiocyanate (PITC) is allowed to react with a free amino residue at the N terminus to prepare a phenylthiocarbamyl derivative (PTC amino acid), which is then released with trifluoroacetic acid in the form of anilinothiazo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| diameter×30 | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com