Chemically inducible cucumber mosaic virus protein expression system
a technology of chemical induction and plant protein, which is applied in the field of plant molecular biology and plant breeding, can solve the problems of low infection efficiency, risk of releasing competent recombinant virus vectors into the environment, and hindering the use of recombinant plant viruses, and achieves neither increased nor decreased raat production. , the effect of increasing the raat level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of the CMViva Expression System 1
[0088]The following materials were used in this example:
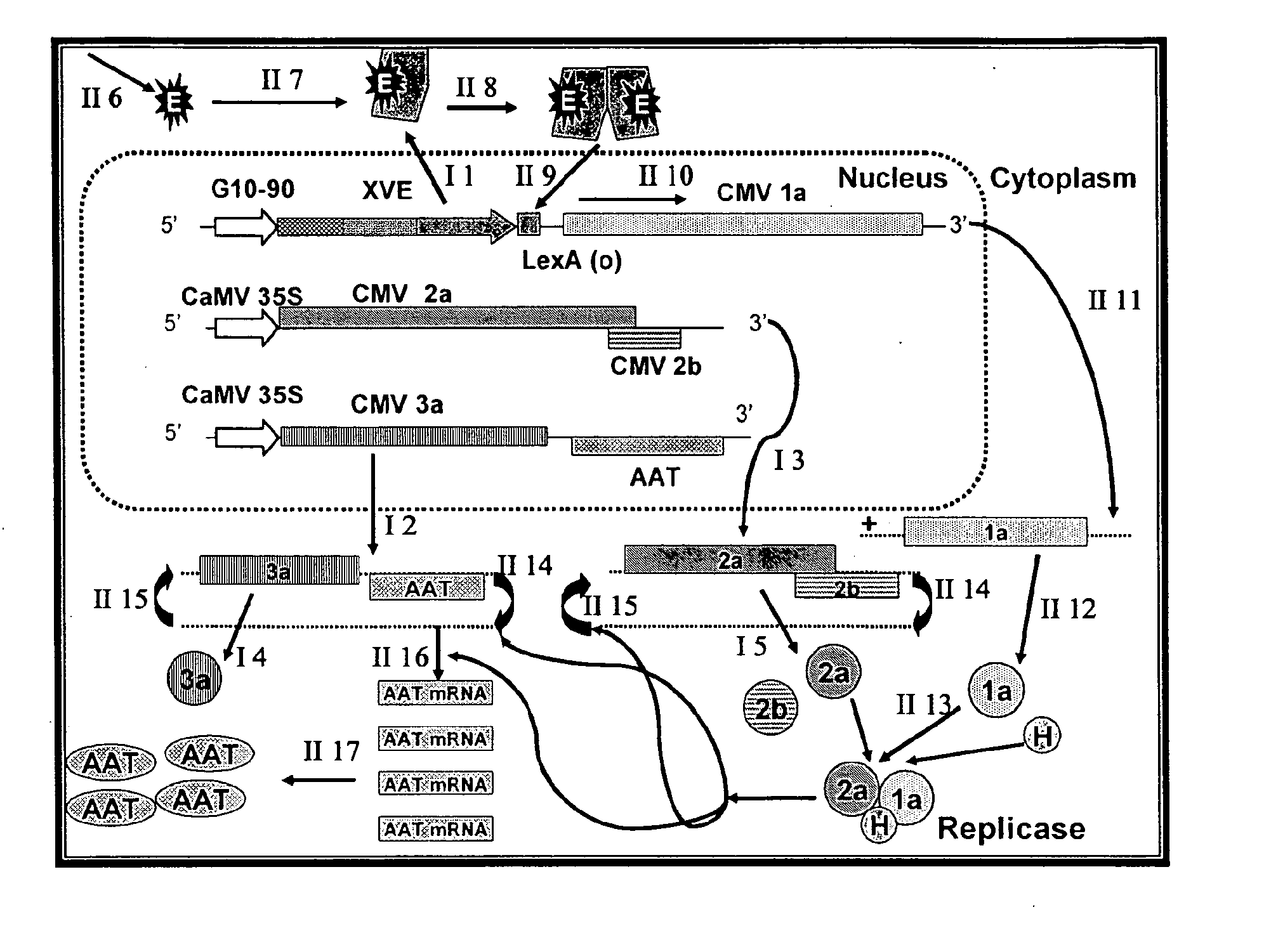
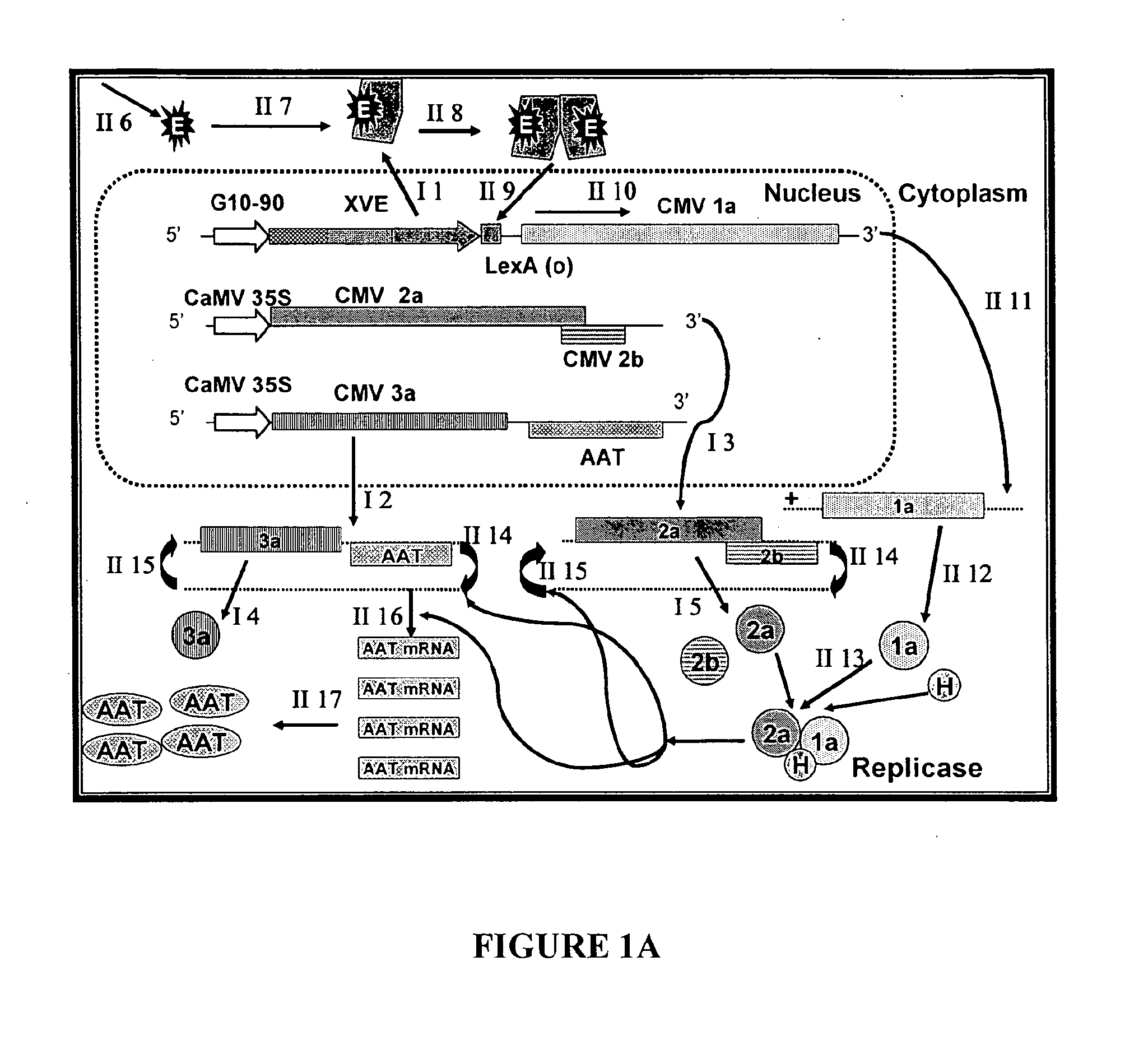
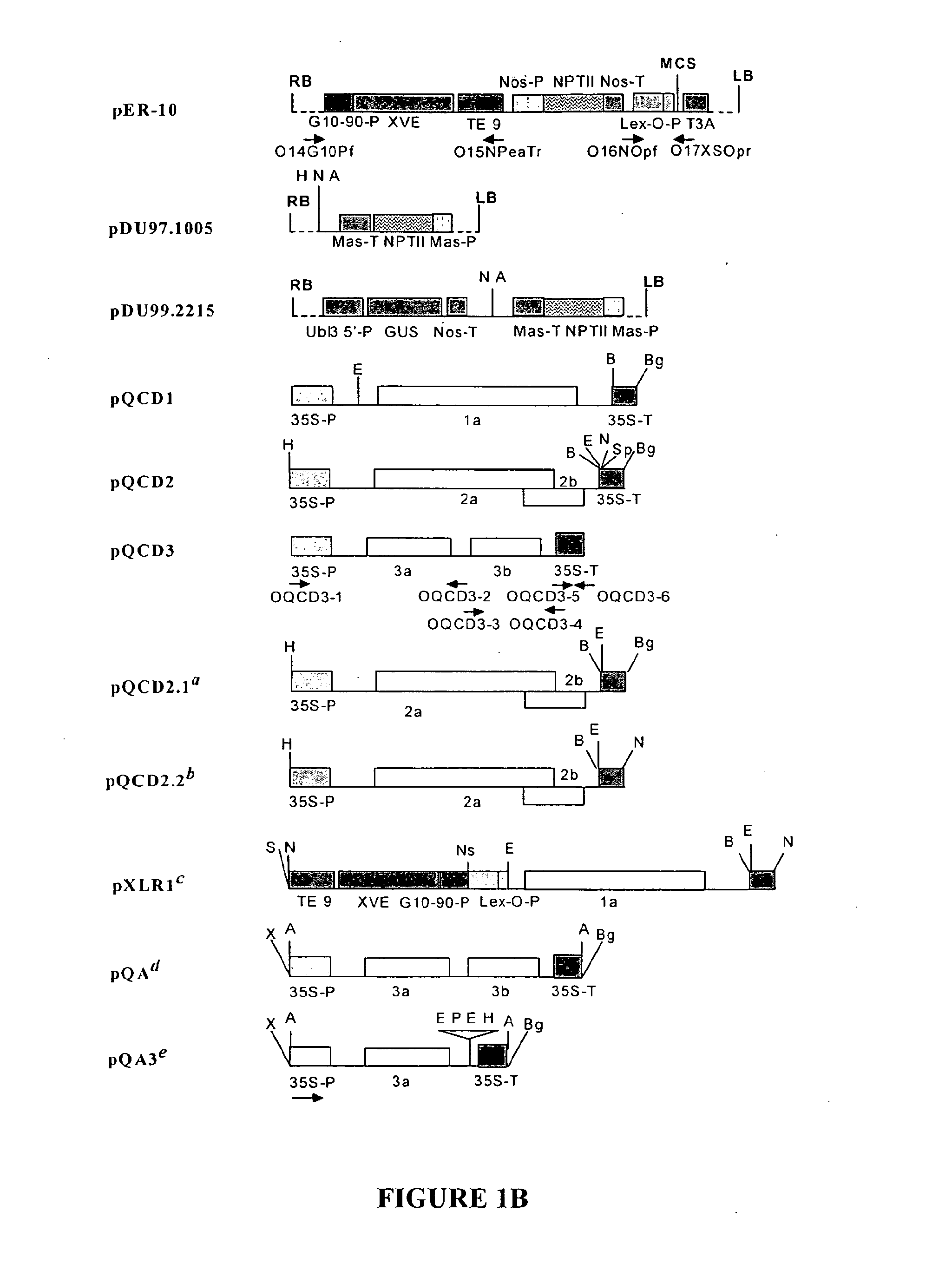
[0089]The complete cDNA clones, pQCD1, pQCD2, and pQCD3, corresponding to genomic RNAs 1, 2 and 3, of CMV strain Q (Ding et al., 1995), respectively.
[0090]The binary plasmid vectors pER8 (Zuo et al., 2000) and pER10 containing the estradiol inducible expression system (XVE system); plasmid pER10 is similar to pER8 except it contains nptII as the selectable marker instead of the hygromycin resistance gene.
[0091]The binary vector p35S:p19 (Voinnet et al., 2003) containing the TBSV p19 gene
[0092]The binary vectors pDU97.1005 (Uratsu and Dandekar, unpublished), a modified version of pCGN1547 (McBride and Summerfelt, 1990)), pDU99.2215 (Escobar et al., 2001) and pART7 (Gleave, 1992).
[0093]A. tumefaciens strain EHA105:pCH32 (Hamilton, 1997) was used to carry all the binary vectors used in this study except for p35S:p19, where A. tumefaciens strain C58C1 was used.
[0094]A series of binary p...
example 2
Construction of the CMViva Expression System 2
[0095]Whereas the CMViva expression system in Example 1 is a binary pCMV-SPAAT plasmid (FIG. 2B), CMViva expression system can also be two separate binary plasmids. The first binary plasmid is the pDUR22XLR1R (FIG. 27); the second binary plasmid is comprised of a series of ten plasmids containing the AscI fragment for pQA (FIG. 28) cloned into pDU97 giving plasmids pQA-2, pQA-4, pQA-6, pQA-7, pQA-8, pQA-9, pQA-10, pQA-11, pQA-12, and pQA-13, collectively called the pQA series hereafter. The pQA series is modified to have restriction sites including, but not limited to, Eco RI-Sac, I-Kpn, 1-Sma, I-BamHI-Xba, I-Acc, I-Sal, I-Pst, and I-SphI-Hind III. The heterologous gene is cloned into the multiple cloning site (MCS) of one or all of the pQA series of plasmids. The different restriction sites used for cloning the recombinant genes allow for easier and more efficient cloning and expression of the heterologous genes, as well as for the iden...
example 3
Agroinfiltration and Induction of Transactivation
[0098]A. tumefaciens EHA105:pCH32 cells containing the appropriate plasmids were grown for 24 to 48 hours in 2 ml LB broth. Approximately 0.5 ml was then transferred to 25 ml LB medium supplemented with 10 μl of 100 mM acetosyringone (3′,5′-dimethoxy-4′-hydroxyacetophenone) (Aldrich Chemicals, Milwaukee, Wis.) and 0.5 ml of MES buffer (pH 5.6) and grown overnight at 28° C. with shaking until cell density (0D600) reached 1.0 absorbance units. Cells were harvested by centrifuging at 2600 g, resuspended in 10 ml sterile de-ionized water, and cell density was adjusted to 1.0 absorbance units. Five ml of the A. tumefaciens cell suspension for each plasmid (p35S-SPAAT, pXVE-SPAAT, and pCMV-SPAAT) was separately mixed with either five ml of sterile water or A. tumefaciens cells containing the p35S:p19 plasmid, then supplemented with magnesium chloride to reach a final concentration of 10 mM and acetosyringone to 150 μM and incubated at room ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com