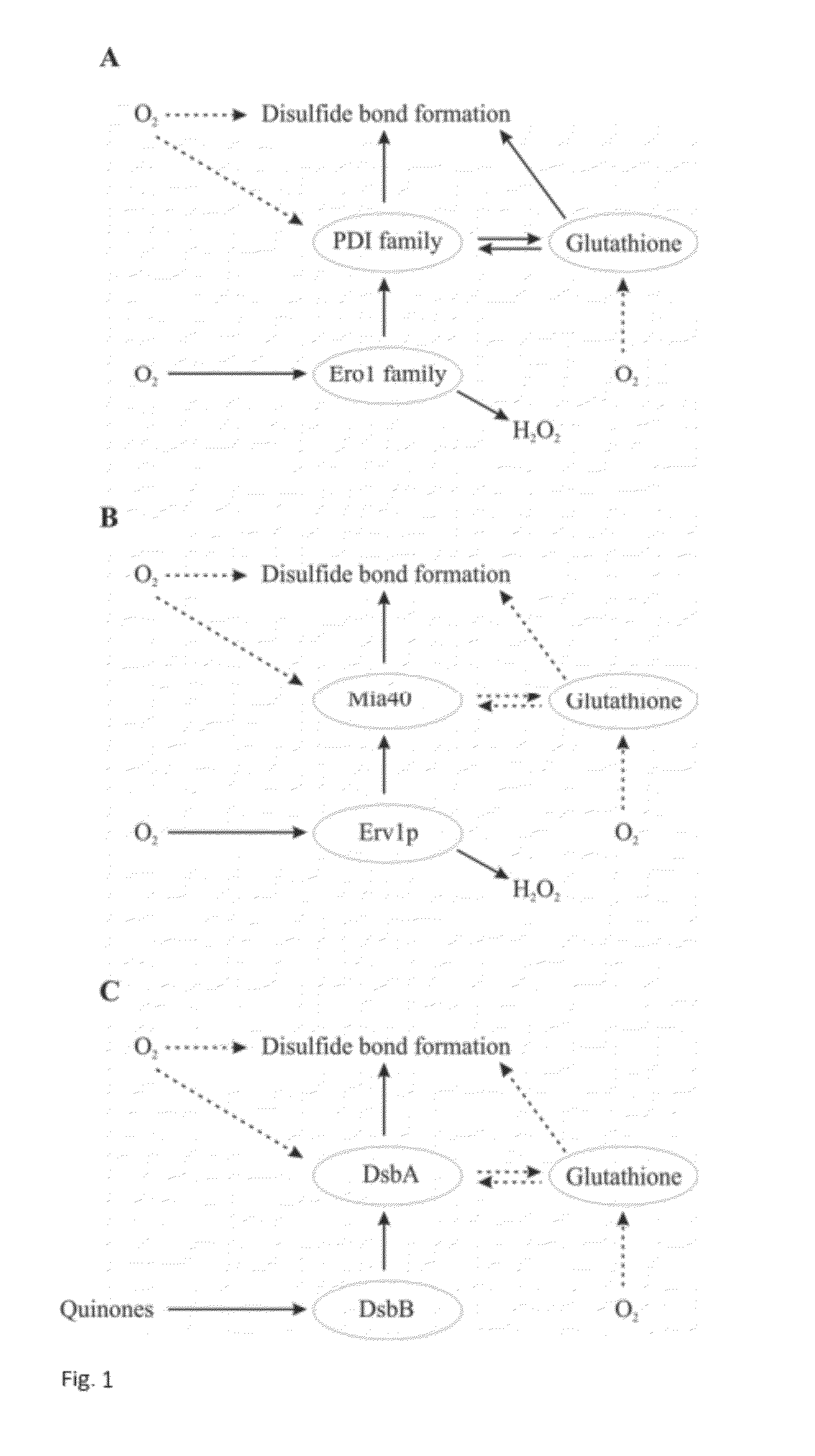
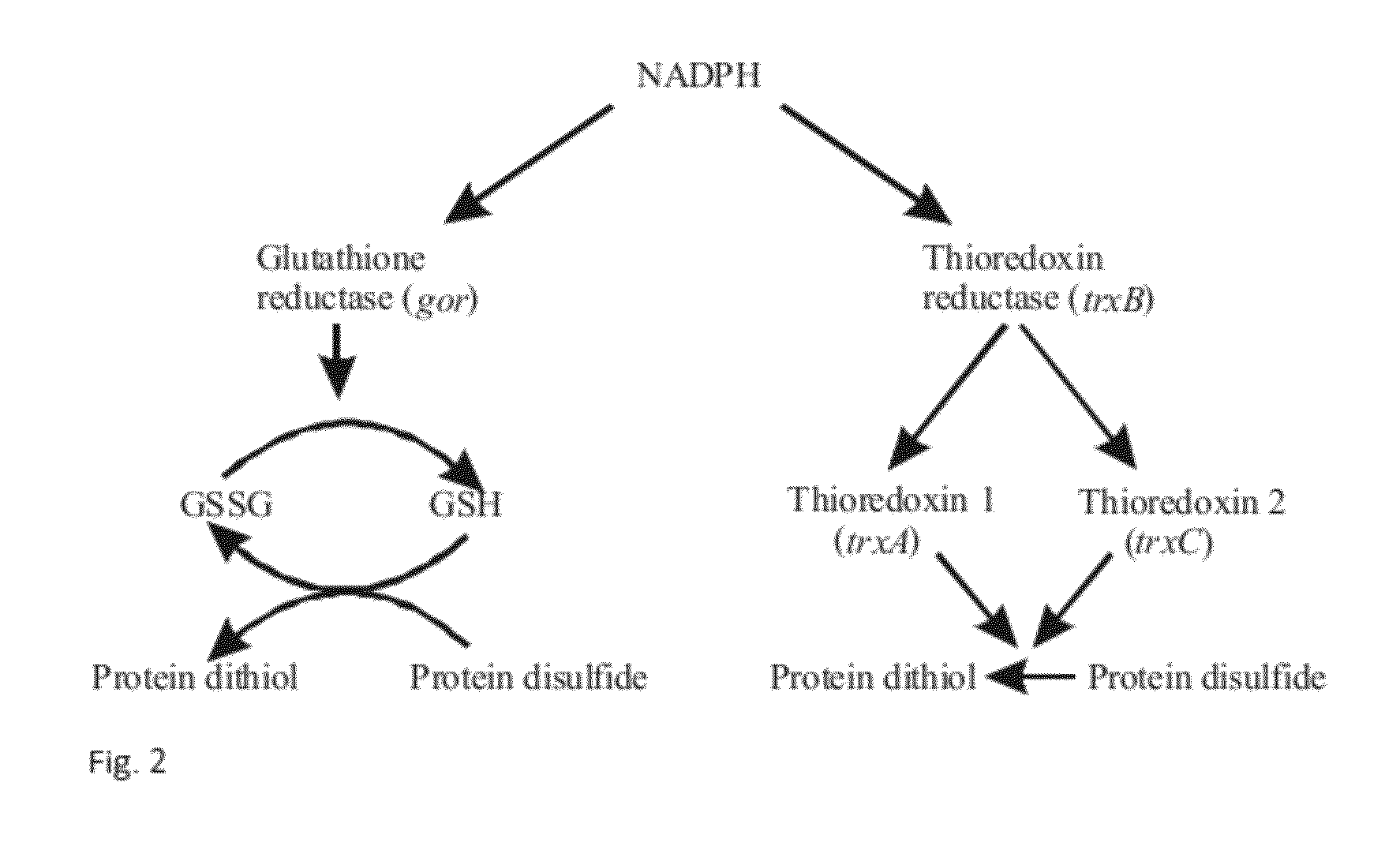
Method for Producing Natively Folded Proteins in a Prokaryotic Host
a prokaryotic host and native folding technology, applied in the field of native folding proteins in prokaryotic hosts, can solve the problems of difficult biotech industry production of proteins containing disulfide bonds on a large scale, the rate-limiting step of native disulfide bond formation, and the confusion of their precise individual roles
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0157]Plasmid constructs used in the protein expressions were generated as part of prestudies (see FIG. 5). The original constructs were polycistronic with multiple genes encoded by a single mRNA driven by a T7 promoter system (IPTG inducible) in a modified version of pET23. These included the protein of interest alone (A), this plus the sulfhydryl oxidase (B) or plus the sulfhydryl oxidase and either PDI or DsbC as a thiol-disulfide isomerase (C).
[0158]Subsequent constructs had the sulfhydryl oxidase (D) or the sulfhydryl oxidase plus the thiol-disulfide isomerase (E) on an arabinose promoter as part of a modified version of pLysS. Constructs D and E are fully compatible with construct A and allow easy inter-conversion of the protein of interest in the system. In addition, constructs D and E allow pre-expression or co-expression of the sulfhydryl oxidase, or the sulfhydryl oxidase and thiol-disulfide isomerase, and the protein of interest.
example 2
Efficient Production of the Luminal Domain of Human Tissue Factor
[0159]Tissue factor (TF), also known as thromboplastin factor III, is a protein involved in the coagulation of blood. It is a transmembrane protein whose luminal domain contains two sequential disulfide bonds.
[0160]pVD81, is a derivative of pET23a which has cloned into the multi-cloning site a gene encoding for an N-terminal hexa-histidine tag (MHHHHHHM) (SEQ ID NO: 11) followed in frame with the luminal domain of human tissue factor (sTF) as represented by the fragment Ser 33-Glu 251 of the full length protein. This vector expresses sTF upon induction with IPTG.
[0161]pVD77 is a derivative of pVD81, in which the gene for the sulfhydryl oxidase Erv1p from S. cerevisiae has been cloned (Met 1-Glu 189) (SEQ ID NO: 5) after the gene for sTF (with suitable ribosome binding sites to initiate translation of both; see FIG. 5). This polycistronic vector co-expresses sTF and Erv1p upon induction with IPTG.
[0162]E. coli strains t...
example 3
Efficient Production of E. coli Alkaline Phosphatase
[0165]Alkaline phosphatase is a hydrolase which removes phosphate groups from many types of molecules. The bacterial enzyme folds in the periplasm. It has two sequential disulfide bonds whose formation is essential for activity. Since it is easily assayed bacterial alkaline phosphatase is widely used as a model protein for disulfide bond formation in vivo.
[0166]pVD80, is a derivative of pET23a which has cloned into the multi-cloning site a gene encoding for an N-terminal hexa-histidine tag (MHHHHHHM) (SEQ ID NO:12) followed in frame with the mature form of E. coli alkaline phosphatase (PhoA) as represented by the fragment Arg 22-Lys 471 of the full length protein. This vector expresses alkaline phosphatase upon induction with IPTG.
[0167]pVD82 is a derivative of pVD80, in which the gene for the sulfhydryl oxidase Erv1p from S. cerevisiae has been cloned (Met 1-Glu 189) (SEQ ID NO: 5) after the gene for E. coli alkaline phosphatase (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com