Methods and Products for Producing Engineered Mammalian Cell Lines With Amplified Transgenes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Targeted Gene Insertion into the CHO DHFR Locus Using Engineered Meganucleases
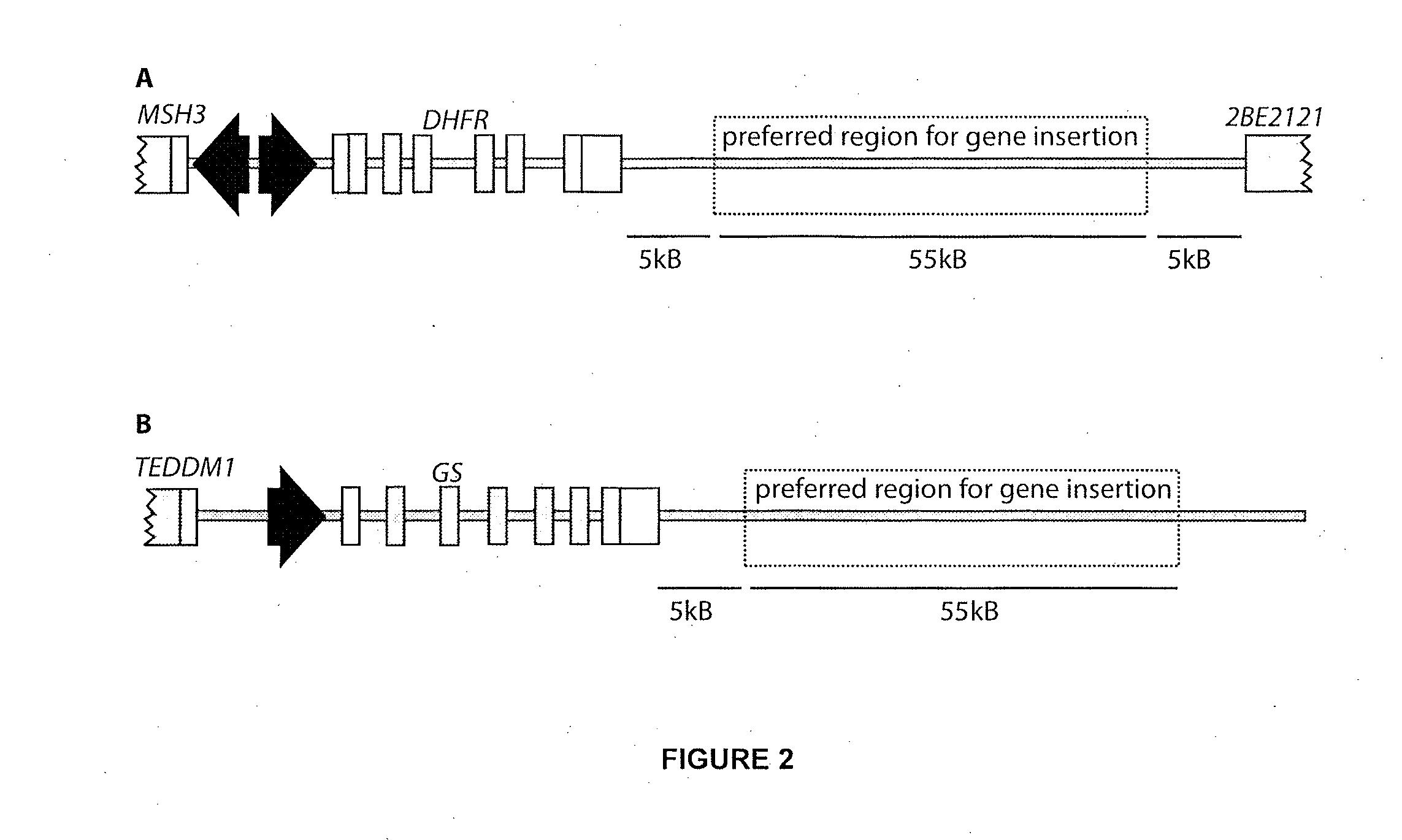
[0136]The CHO genomic DNA sequence 10,000-55,000 base pairs downstream of the DHFR gene was searched to identify DNA sites amenable to targeting with engineered meganucleases. Two sites (SEQ ID NO: 7 and SEQ ID NO: 8) were selected which are, respectively, 35,699 and 15,898 base pairs downstream of the DHFR coding sequence (Table 2).
TABLE 2Example Recognition Sites For EngineeredMeganucleases in the CHO DHFR Locus.SEQLocation RelativeIDto CHO DHFRNO:Target Site SequencesCoding Sequence75′-TAAGGCCTCATATGAAAATATA-3′35,699 bp downstream85′-ATAGATGTCTTGCATACTCTAG-3′15,898 bp downstream
1. Meganucleases that Recognize SEQ ID NO: 7 and SEQ ID NO: 8
[0137]An engineered meganuclease (SEQ ID NO: 9) was produced which recognizes and cleaves SEQ ID NO: 7. This meganuclease is called “CHO-23 / 24”. A second engineered meganuclease (SEQ ID NO: 10) was produced which recognizes and cleaves SEQ ID NO: 8. This meganuclease is...
example 2
Insertion of an Engineered Target Sequence into the CHO DHFR or GS Gene Coding Regions
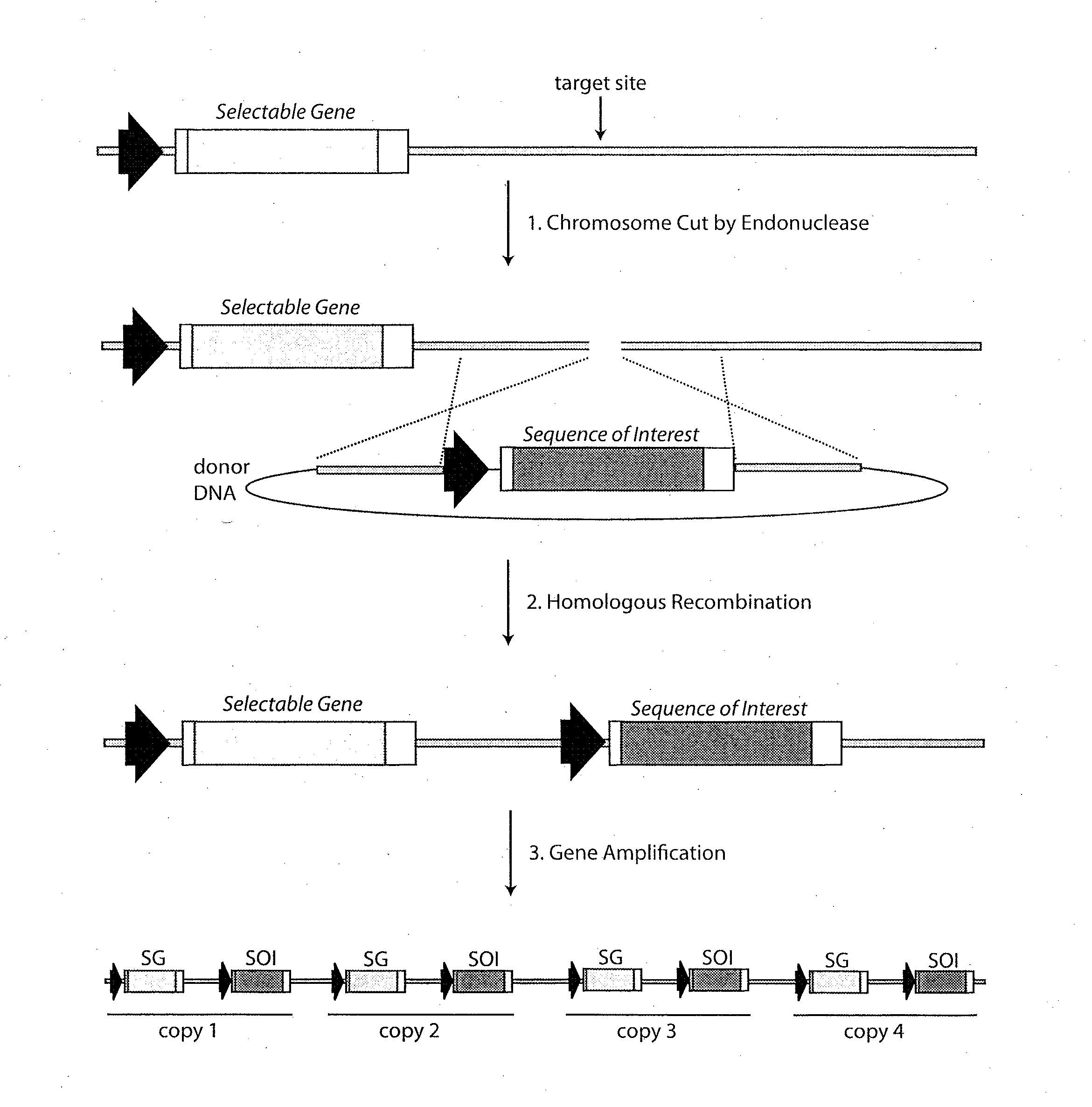
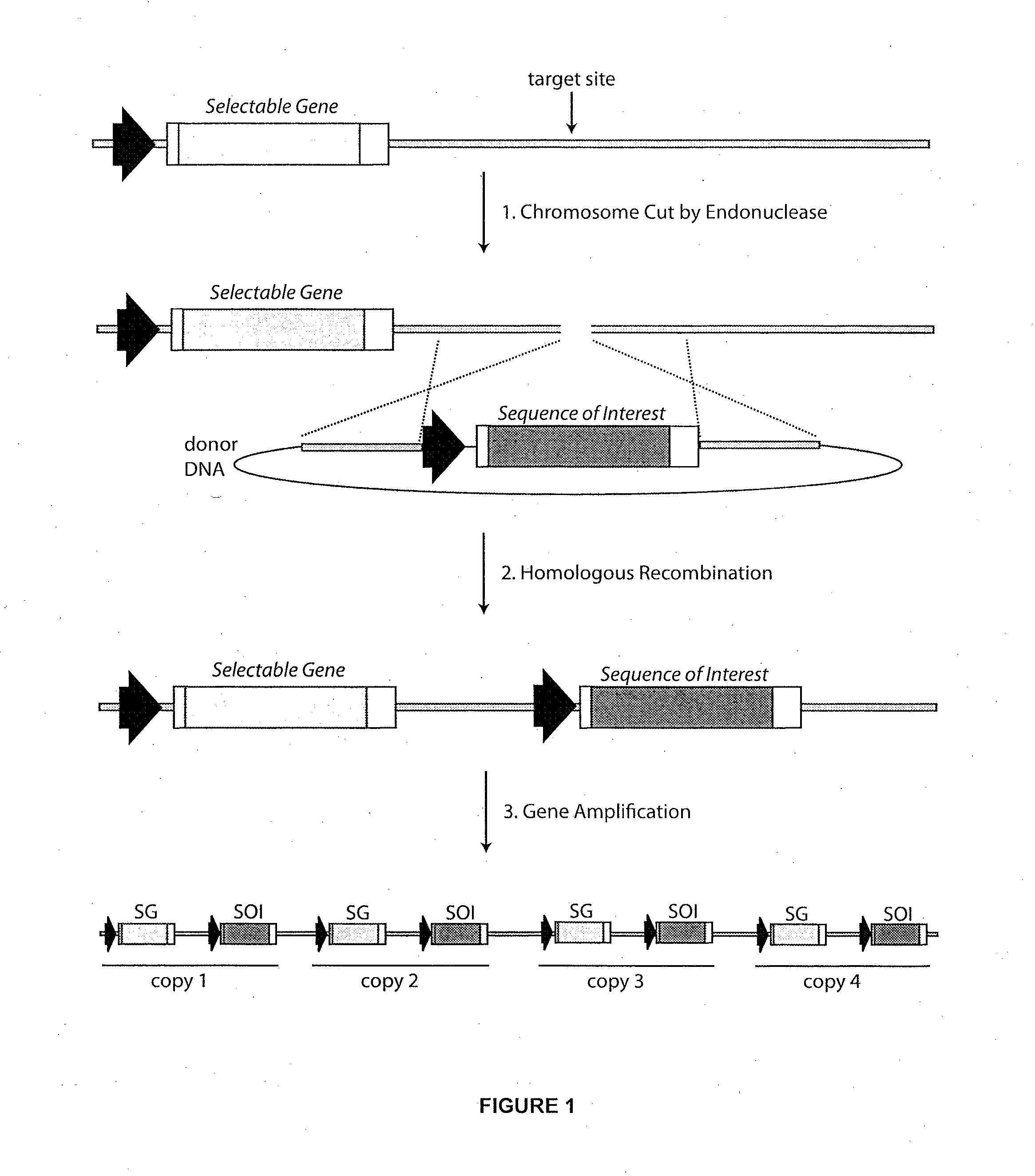
[0150]As diagrammed in FIG. 4, an alternative method for targeting a sequence of interest to an amplifiable locus involves the production of a cell line in which a portion of a selectable gene is replaced by an engineered target sequence. The advantage of this approach is that the subsequent insertion of a sequence of interest can be coupled with reconstitution of the selectable gene so that cell lines harboring the properly targeted sequence of interest can be selected using the appropriate media conditions. A cell line harboring such an engineered target sequence can be produced using nuclease-induced homologous recombination. In this case, a site-specific endonuclease which cuts a recognition sequence near or within the selectable gene sequence is preferred.
1. Engineered Meganucleases that Cut within the DHFR or GS Genes.
[0151]A meganuclease called “CHO-13 / 14” (SEQ ID NO: 12) was produced which ...
example 3
Meganucleases for Targeting Gene Insertion to the CHO GS Locus
[0155]1. Engineered Meganucleases that Cut Downstream of the CHO GS Gene.
[0156]An engineered meganuclease called “CHOX-45 / 46” (SEQ ID NO: 29) was produced which recognizes a DNA sequence (SEQ ID NO: 30) approximately 7700 base pairs downstream of the CHO GS coding sequence. CHO cells were transfected with mRNA encoding CHOX-45 / 46 as described in Example 2. 72 hours post transfection, genomic DNA was extracted from the transfected cell pool and the region downstream of the CHO GS gene was PCR amplified using a pair of primers (SEQ ID NO: 31 and SEQ ID NO: 32) flanking the CHOX-45 / 46 recognition sequence. PCR products were then cloned and 24 cloned products were sequenced. It was found that 14 of the 24 clones PCR products (58.3%) had large mutations in the sequence consistent with meganuclease-induced genome cleavage followed by mutagenic repair by non-homologous end-joining From these data, we conclude that the CHOX-45 / 46...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com