Histone demethylation mediated by the nuclear amine oxidase homolog LSD1
a nuclear amine oxidase and histone demethylation technology, applied in the field of gene regulation, can solve the problems of unclear whether bona fide histone demethylases exist, the molecular identity of these putative histone demethylases remains elusive, and cannot allow dynamic regulation of histone methylation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0115]To understand the function and mechanism of action of KIAA0601, we undertook molecular, biochemical and enzymological analyses of the protein. Using multiple experimental approaches, we demonstrate that KIAA0601 is a lysine-specific demethylase with substrate specificity for K4 methylated histone H3. We now refer to protein as LSD1 (Lysine Specific Demethylase 1) to reflect this newly identified role. The text and figures corresponding to this example may be found in Shi et al. Cell (2004) 119:903, which is specifically incorporated by reference herein.
[0116]LSD1 is a Transcriptional Co-repressor that is Evolutionarily Conserved
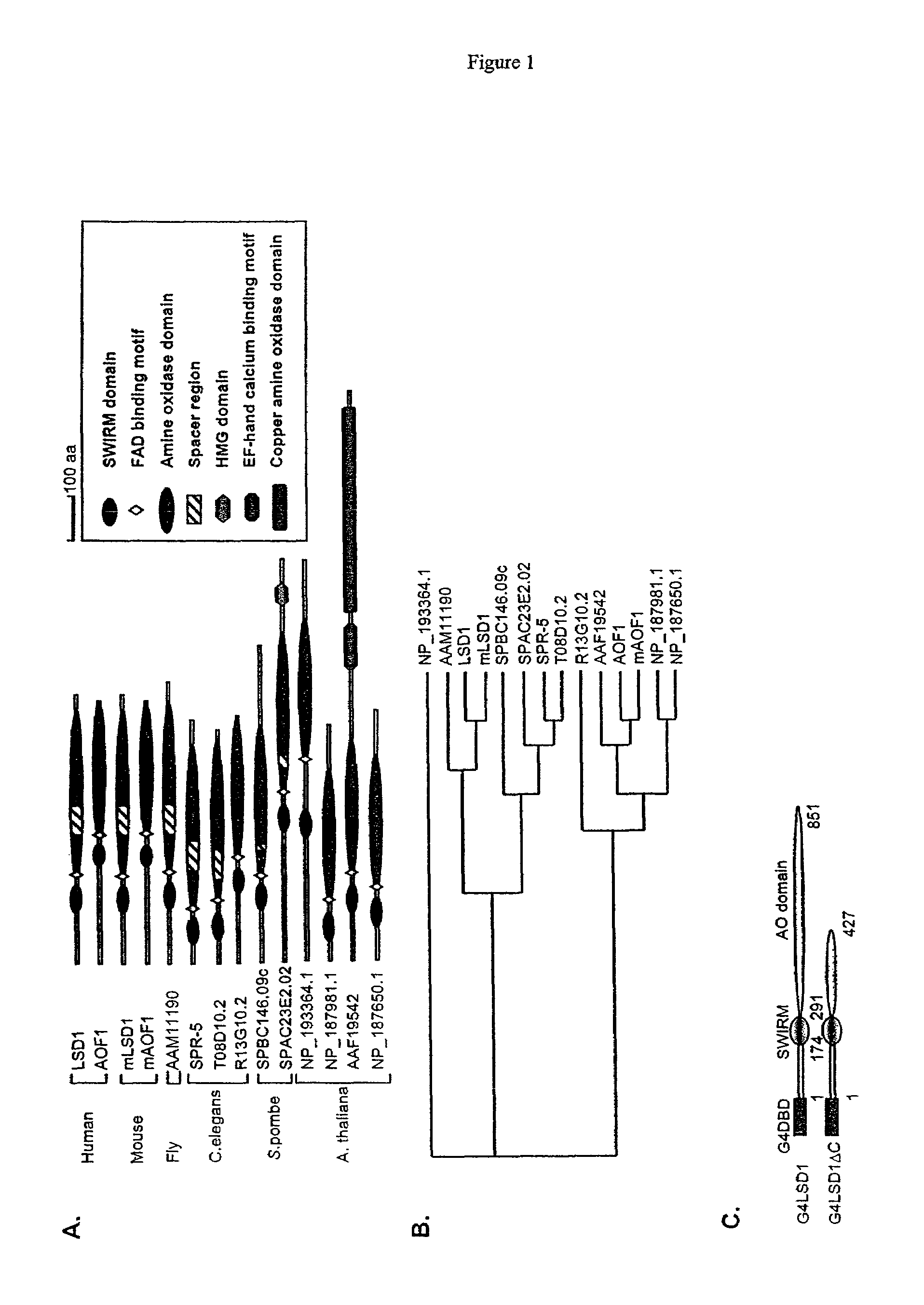
[0117]FIG. 1A shows a schematic diagram of the predicted domains of LSD1 and its related proteins. The C-terminal 2 / 3 of LSD1 display significant sequence homology with FAD-dependent amine oxidases. The N-terminus of LSD1 has a SWIRM domain, which is found in a number of proteins involved in chromatin regulation (Aravind and Iyer, 2002). Although the fu...
example 2
[0119]LSD1 is a Lysine-specific Histone Demethylase
[0120]LSD1 is a flavin-containing protein based on its ability to bind FAD ((Humphrey et al., 2001), and data not shown). Its sequence homology with amine oxidases predicts that LSD1 may catalyze oxidation reactions of biogenic amines including monoamine, polyamines or N-methylated protein substrates (such as histones) (Bannister et al., 2002). Amine oxidation catalyzed by flavin-containing amine oxidase is characterized by oxidative cleavage of the α-carbon bond of the substrate to form an imine intermediate, which, in turn, is hydrolyzed to form an aldehyde and amine via a non-enzymatic process. In a complete catalytic cycle, the cofactor FAD is reduced to FADH2 and then is likely to be re-oxidized by oxygen to produce hydrogen peroxide (Binda et al., 2002). We hypothesized that, as a flavin-containing amine oxidase homolog, LSD1 may catalyze the conversion of mono- or dimethylated K (or R) to non-methylated K (or R) and formaldeh...
example 3
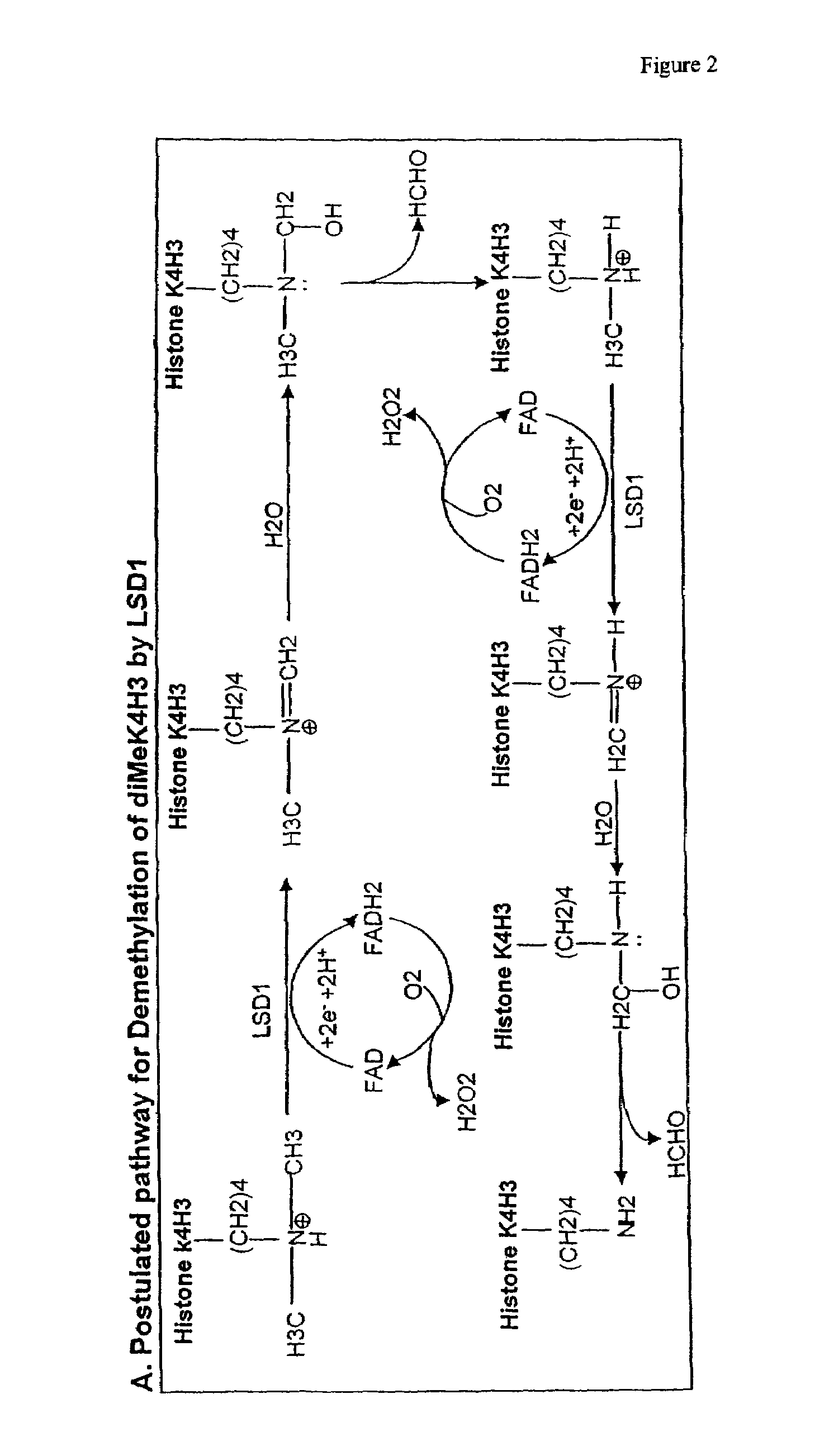
[0124]LSD1-mediated Histone Demethylation Generates Formaldehyde
[0125]We used a third independent method to investigate the possibility that LSD1 is a histone demethylase. As shown in FIG. 2, the demethylation reaction mediated by LSD1 is predicted to generate formaldehyde. To determine whether formaldehyde was produced in LSD1-mediated enzymatic reactions, we first used the formaldehyde dehydrogenase (FDH) assay to detect the presence of formaldehyde (Lizcano et al., 2000). This assay employs formaldehyde dehydrogenase to convert formaldehyde to formic acid using NAD+ as the electron acceptor, whose reduction to NADH can be spectrophotometrically measured at OD 340 nm. Thus, when the demethylation reaction is coupled with the FDH assay, the enzymatic activity of LSD1 and reaction kinetics can be determined by measuring the production of NADH. A standard curve was first generated using purified FDH (EC 1.2.1.46), NAD+ and different concentrations of formaldehyde ranging from 1 μM to...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| molecular mass | aaaaa | aaaaa |
| molecular mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com