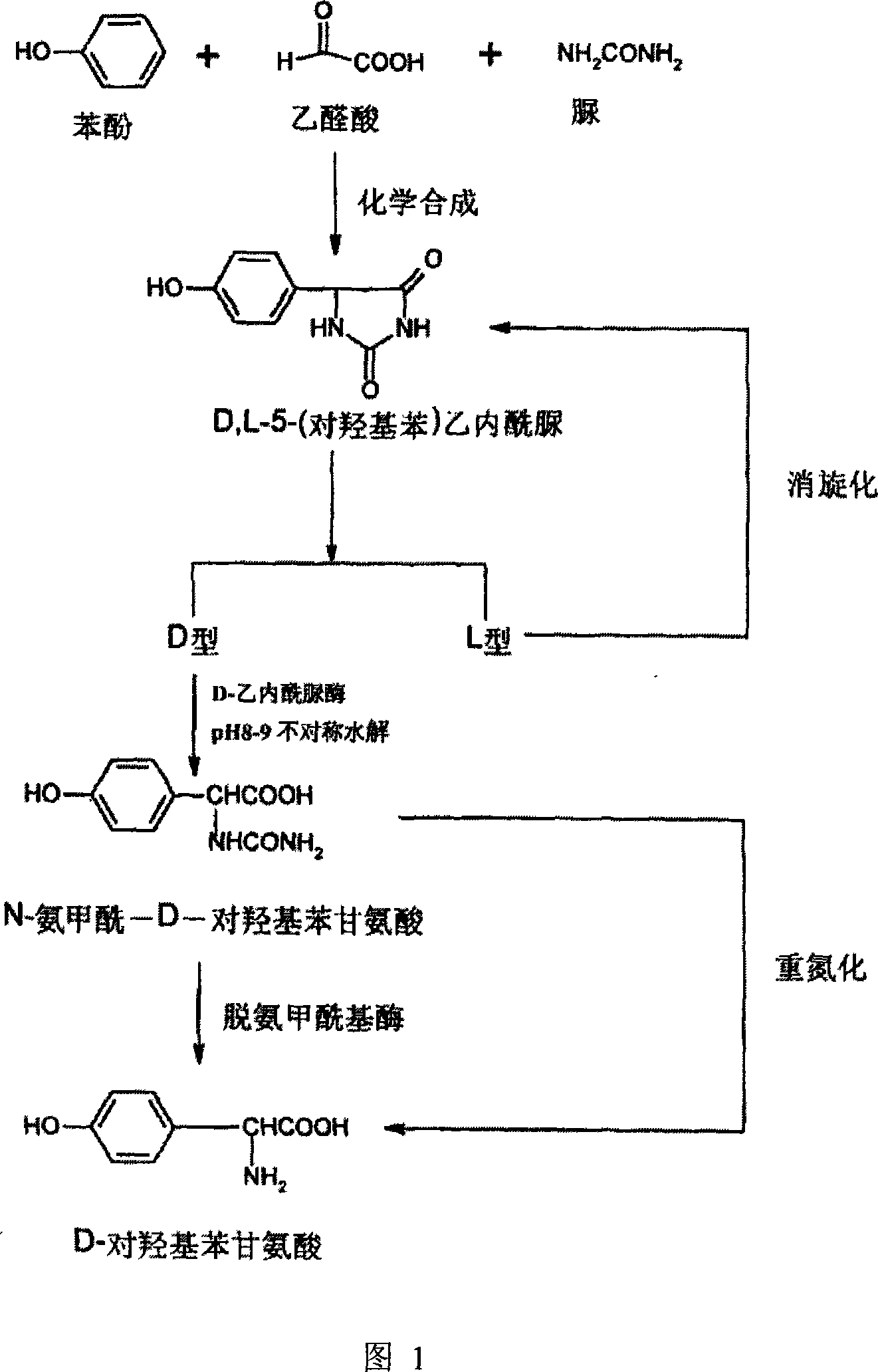
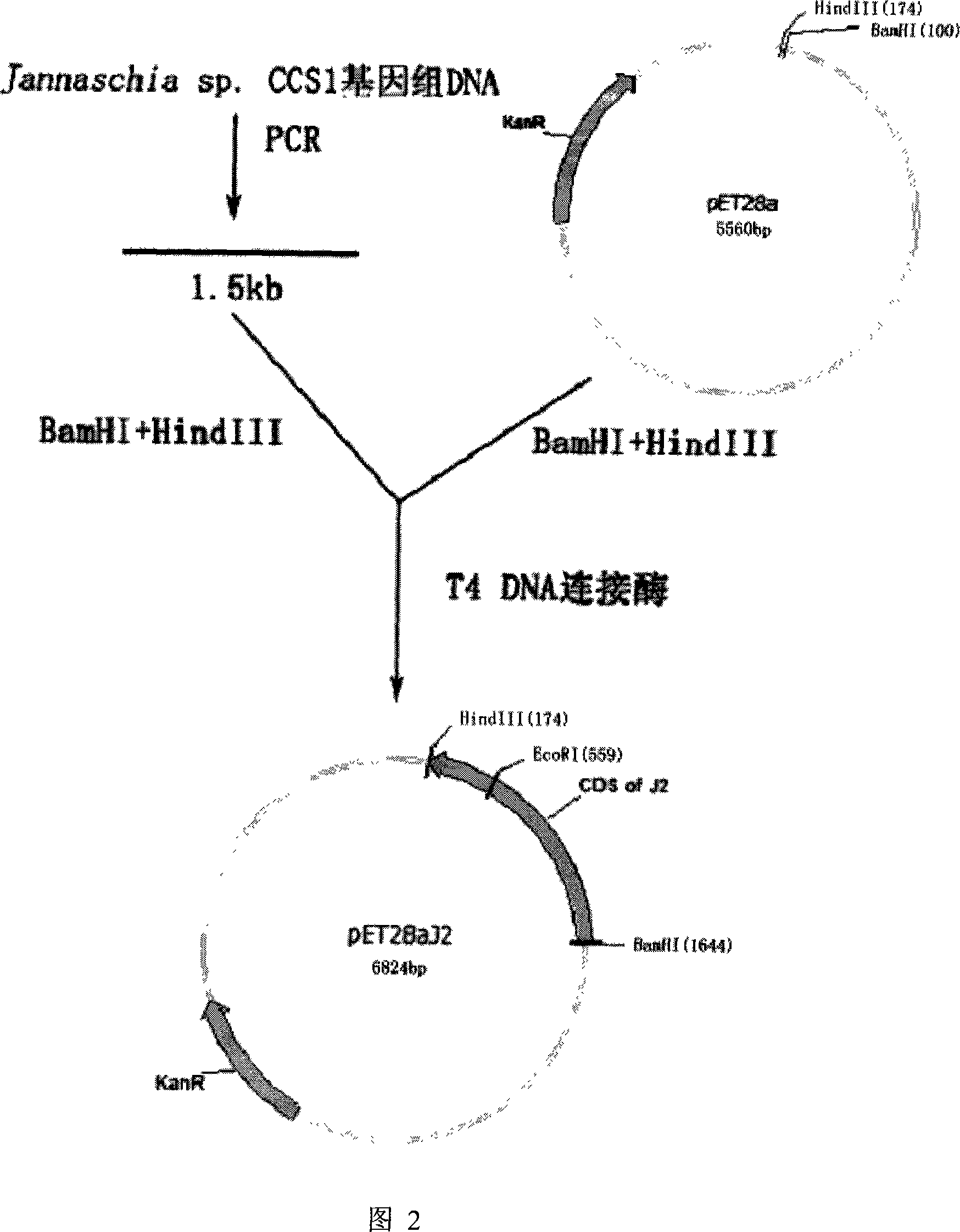
Hydantoinase gene, coded amino acid and application thereof
A technology of gene encoding and hydantoinase, which is applied in the field of genetic engineering, can solve problems such as hydantoinase from cloning sources that have not yet been seen.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1, the construction that contains the expression vector of target gene and engineering bacterium
[0024] 1.1. Primer design
[0025] According to the genome sequence of Jannaschia sp.CCS1 published by Genebank, primers A and B were designed, and their sequences are as follows:
[0026] Primer A: 5'-AGGGGGATCCATGAGCAAGGTGATCAAGGG-3';
[0027] Primer B: 5'-CTAGAAGCTTTCAAACCCCCGCCGGAATG-3'.
[0028] 1.2. PCR amplification
[0029] Use A and B as primers, and use genomic DNA from Jannaschia sp.CCS1 as a template to amplify the target gene by PCR.
[0030] Reaction system: 1×PCR Buffer for KOD plus, 2mM MgSO 4 , 0.4 μM each of primer A and primer B, 100 ng of dJannaschia sp.CCS1 genomic DNA, 0.2 mM dNTP, 1U KOD plus / 50 μl.
[0031] Reaction conditions: 94°C for 5min, 94°C for 30s, 55°C for 30s, 72°C for 1.5min, 30 cycles; 72°C for 10min.
[0032] After the reaction, according to the operation manual, the PCR product was recovered and purified with the glue ...
Embodiment 2
[0045] Embodiment 2, the expression of target gene
[0046] A single colony of E.coli BL21(DE3) / pETJ2 was picked and inserted into LB liquid medium containing 50 μg / ml kanamycin, cultured overnight at 37° C. and 200 rpm.
[0047] Inoculate the overnight cultured BL21(DE3) / pETJ2 into LB liquid medium containing 50 μg / ml kanamycin at 1% inoculum size, cultivate at 37°C, 200 rpm until OD600≈0.6, and add a final concentration of 0.25 mM IPTG, induced culture at 20°C for 5 hours.
[0048] After centrifuging the bacterial solution at 12000rpm for 2 minutes, discard the supernatant.
[0049] The pellet was resuspended in 50 mM Tris-HCl buffer pH 8.0 and sonicated. The broken cells were analyzed by polyacrylamide gel electrophoresis for the expression level of the target protein, and the results are shown in FIG. 3 .
[0050] According to the results in Figure 3, it can be seen that the expression level of the target protein accounts for about 30% of the total protein, and J2 can b...
Embodiment 3
[0051] Embodiment 3, the mensuration of target protease activity
[0052] Insert the BL21(DE3) / pETJ2 cultured overnight in Example 2 into TB liquid medium containing 50 μg / ml kanamycin at an inoculum size of 1%, at 37°C, 200rpm, and cultivate to OD600≈0.6, add IPTG with a final concentration of 0.25 mM was induced at 20° C. for 5 hours.
[0053] Collect the bacterial solution, centrifuge, discard the supernatant, freeze and thaw once at -20°C, and then resuspend the bacterial solution with 50mM Tris-HCl buffer solution with pH 8.0.
[0054] The resuspended cells were added to Tris-HCl buffer solution containing 1% p-hydroxyphenylhydantoin, pH 8.0, reacted at 40°C for 10 minutes, added 3.7% HCl to stop the enzyme reaction and mixed well.
[0055] The above reaction solution was centrifuged at 12000rpm for 5 minutes, and the supernatant was diluted 4 times and then analyzed by HPLC to analyze the amount of reaction product generated (the results are shown in Figure 4), and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com