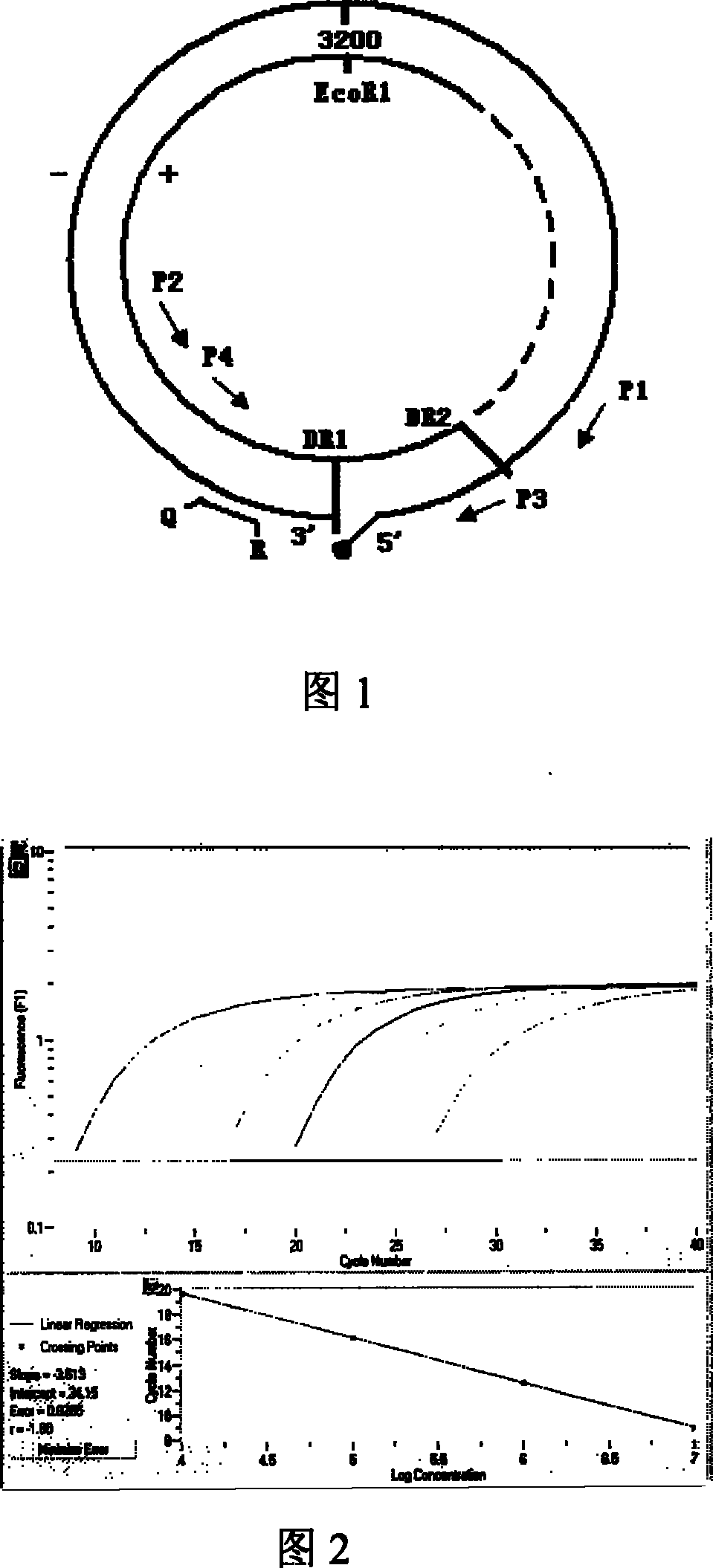
Nest type-real time quantitative PCR method for detecting hepatitis B virus cccDNA
A hepatitis B virus, real-time quantitative technology, applied in biochemical equipment and methods, microbial determination/inspection, etc. Sensitivity, wide application range, accurate results of operation process and result analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0036] Below by example, the present invention is further explained.
[0037] 1. Specimen collection: extract 3ml of fresh whole blood from patients with hepatitis B, and separate serum; extract 5ml of fresh whole blood from patients with hepatitis B, separate PBMCs by density gradient centrifugation, and count cells; collect liver biopsy and liver resection specimens from patients with chronic hepatitis B, cirrhosis, and liver cancer. Freeze at -70°C.
[0038] 2. DNA extraction:
[0039] A. Serum viral DNA was extracted with QIAamp MinElute Virus Spin Kit from QIAGEN;
[0040] B. Plasmid kit to extract virus DNA in PBMC: Plasmid extraction kit (Beijing Tianwei Times Company) was used to extract virus DNA in PBMC.
[0041] C. DNA in liver tissue was extracted with genomic DNA extraction reagent (Beijing Tianwei Times Company).
[0042] The extracted DNA was dissolved in 50 μl TE or deionized water and frozen for future use.
[0043] 3. Design and synthesis of primers and p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com