Salt-endurance related rice protein kinase gene clone and uses thereof
A technology of protein and salt tolerance, applied in application, genetic engineering, plant genetic improvement, etc., can solve problems such as unclear MAPK cascade, achieve the effect of improving tolerance and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1. Acquisition of Salt Tolerance Gene OsMAP3Kβ3
[0048] 1. Acquisition of gene sequence
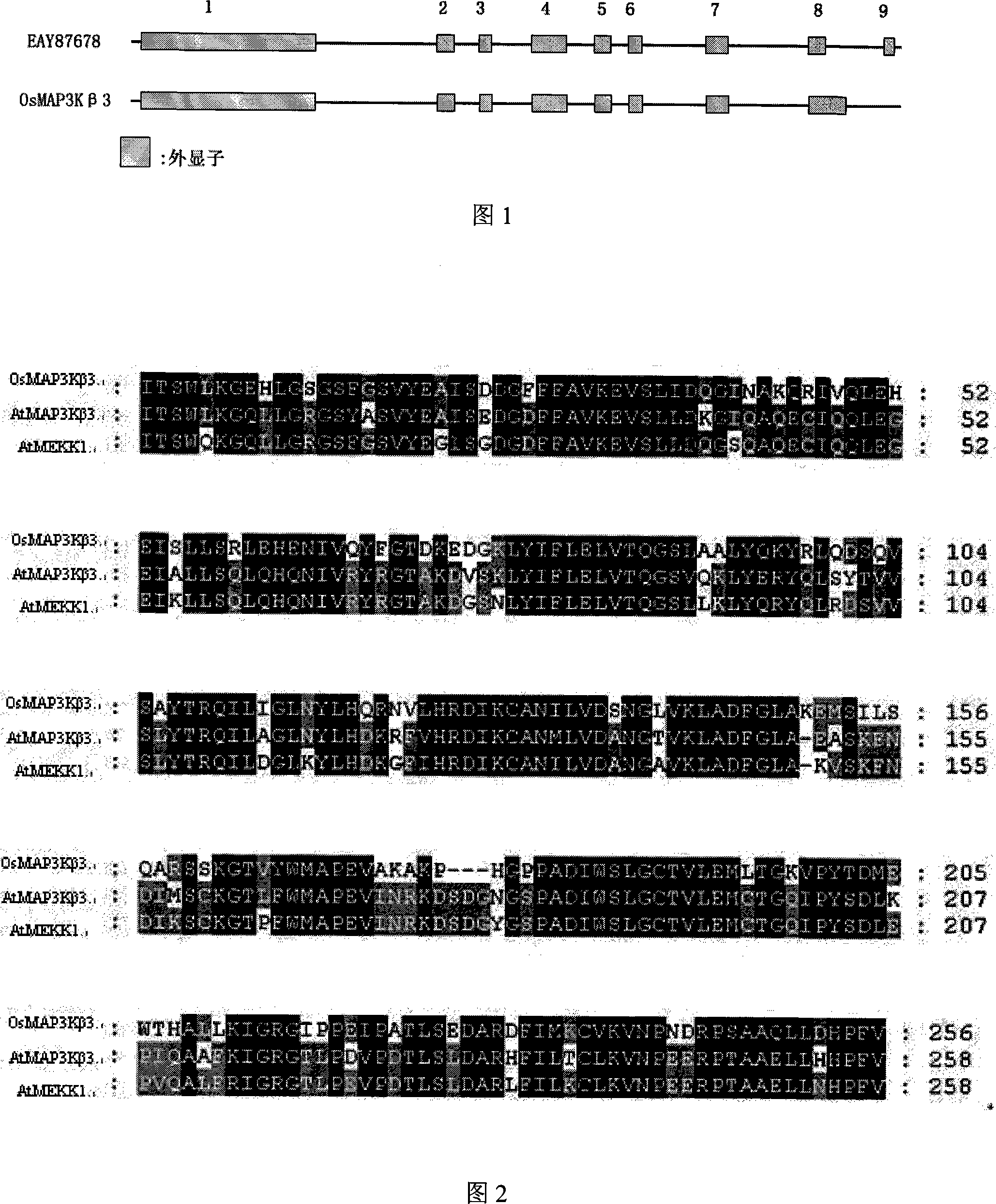
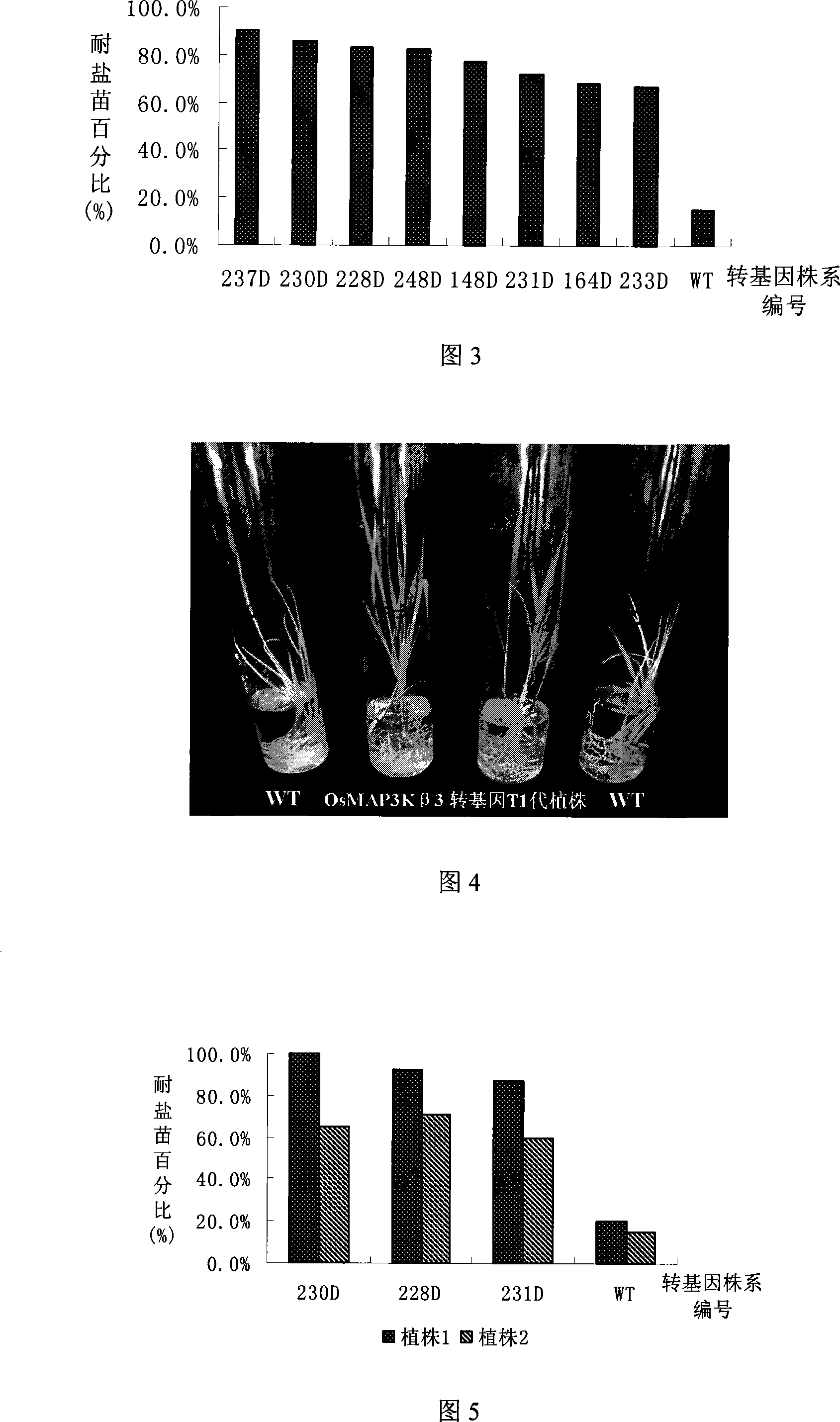
[0049] Using the nucleotide sequence of Arabidopsis MAP3K beta 3 gene (GenBank No.: AJ010092) to search for homologous genes in GENBANK, the homologous sequence of rice (GenBank No.: EAY87678) was obtained. The homologous sequence is located in the rice On the two chromosomes, its CDS is 1572bp, encodes 523 amino acids, has 58% homology with the kinase domain of AtMAP3Kβ3, and has 11 subdomains of protein kinases. It is speculated that it is a MAP3K gene in rice and named is OsMAP3Kβ3.
[0050] 2. PCR amplification and sequence analysis of OsMAP3Kβ3
[0051] Primers were designed according to the coding region of OsMAP3Kβ3 sequence in GENBANK, and OsMAP3Kβ3 was amplified by PCR method using the rice Guanglu dwarf cDNA as a template. The primer sequences are as follows:
[0052] Forward primer: 5'>CACCATGGCGGCGCGCCAGCGG>3;
[0053] Reverse primer: 5'>CCGCTGTATCATGGCAAG...
Embodiment 2
[0056] Example 2, Obtaining of transgenic rice of OsMAP3Kβ3 gene related to salt tolerance in rice
[0057] Using the Agrobacterium-mediated method to transform rice with the gene OsMAP3Kβ3 related to salt tolerance obtained in Example 1, the specific method is as follows:
[0058] 1) Transformation of Agrobacterium
[0059] The OsMAP3Kβ3 gene in the recombinant plasmid pENTER-TOPO Vector-OsMAP3Kβ3 constructed in Example 1 was recombined into the expression vector pH2GW7 (Gateway of Invitrogen Company) by LR reaction TW vector vector), the LR reaction system is 2 μl buffer, 2 μl (150-300ng) linearized pH2GW7 plasmid DNA, 2 μl (100-300ng) pENTER-TOPO Vector-OsMAP3Kβ3 plasmid DNA, 2 μl H 2 O, then add 2 μl LRClonase, the LR reaction condition is 25°C water bath for 1 hour, add 1 μl 2U / μl proteinase K, 37°C water bath for 10 minutes. Then, the above-mentioned recombinant vector was transformed into Escherichia coli (E.coli) DH5α competent cells by the heat shock method, positi...
Embodiment 3
[0064] Example 3, Genetic Analysis and Molecular Identification of Transgenic T1 Generation Rice Plants
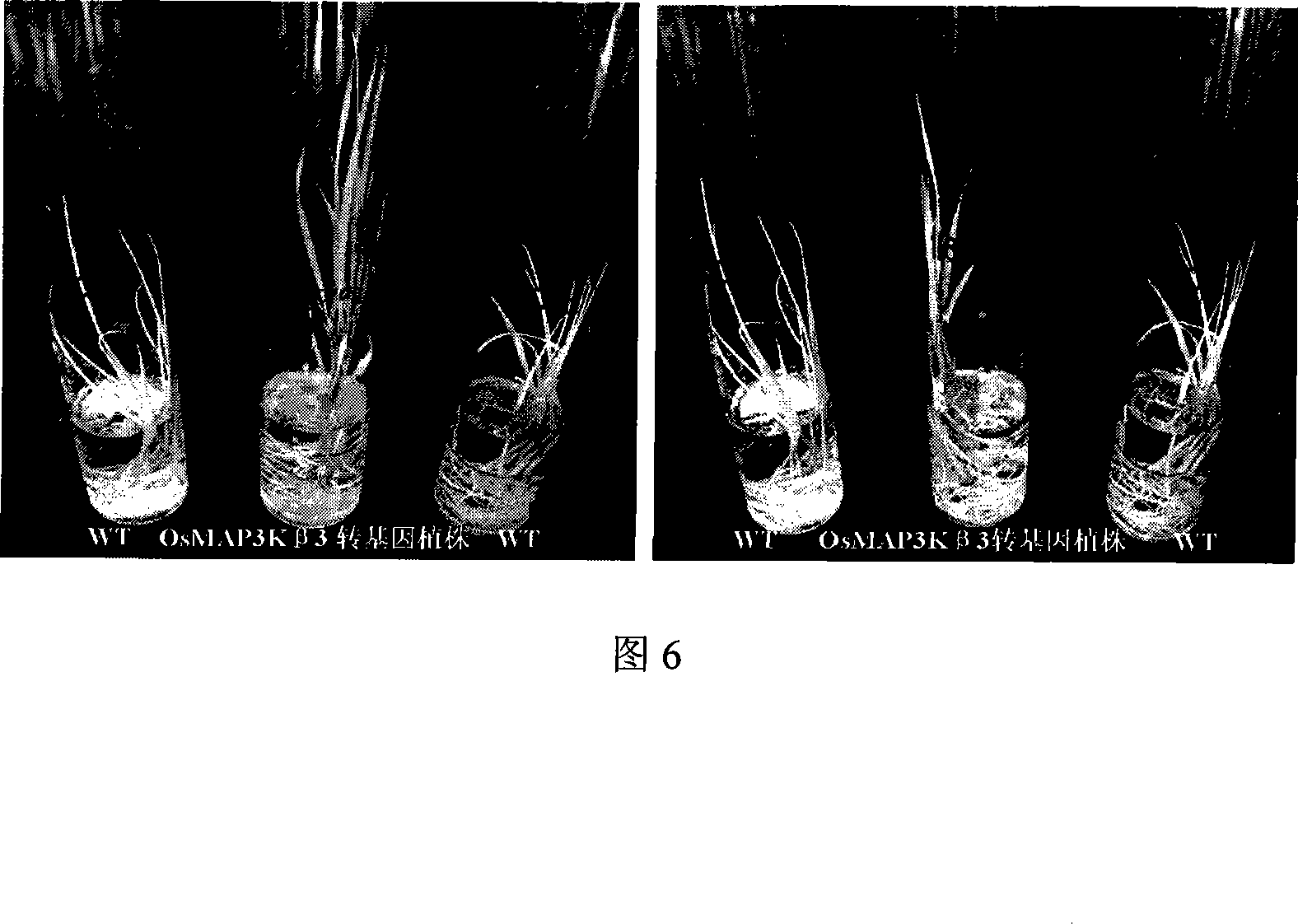
[0065] The seeds of the OsMAP3Kβ3 T1 generation transgenic rice harvested in Example 2 were soaked in water for 3 days to allow them to germinate, and then 50 mg / L hygromycin was used for resistance selection, and at the same time, non-transgenic rice Zhonghua 11 was used as a control. After 5 days of selection, All the control plants died, counted the number of resistant seedlings and dead seedlings of the transgenic plants, analyzed the resistant segregation ratio of the transgenic plants, and selected lines with a segregation ratio of resistant seedlings and non-resistant seedlings of about 3:1, the results showed that OsMAP3Kβ3 transgenic lines with a single site insertion.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com