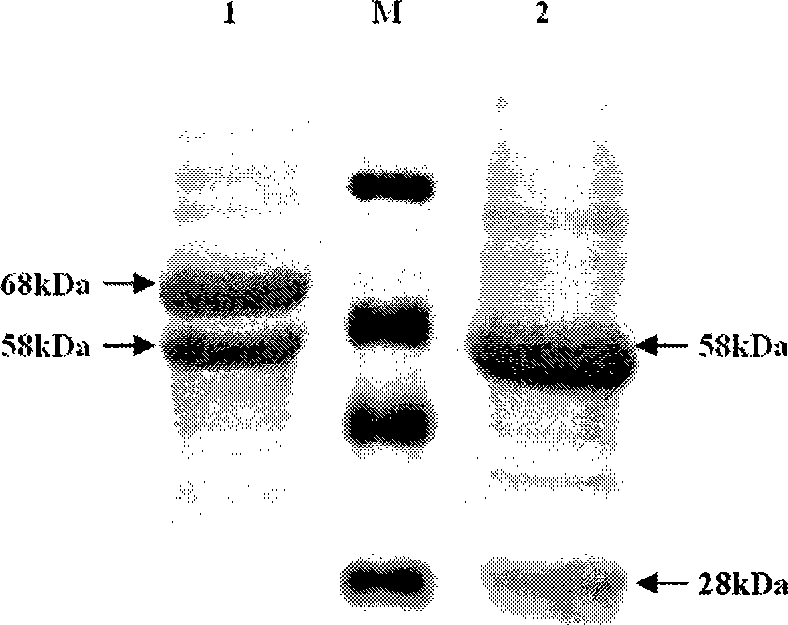
Cephalosporin C acrylase and its vector and application
A technology of acylase and acylase activity, applied in the field of bioengineering, can solve problems such as inability to catalyze CPC substrates, and achieve the effects of high expression activity, high product tolerance, and high catalytic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Design and Synthesis of Cephalosporin C Acylase Gene Sequence
[0044] (1) CPC acylase gene design of the present invention: the protein sequence of Pseudomonas sp.SE83cephalosporin acylase II in Genebank (SEQ ID NO: 1, Genbank AAA25690) was used, and six mutation sites in SEQ ID NO: 3 were introduced at the same time: Val122Ala-Gly140Ser-Phe58βArg-Ile75βThr-Ile176βVal-Ser471βCys (WO2005 / 014821A1), using the online server DNAWorks (URL: http: / / helixweb.nih.gov / dnaworks) to reverse design the gene sequence of CPC acylase. According to the codon degeneracy and preference of E. coli, the above-mentioned CPC acylase gene sequence redesigned for high expression in E. coli hosts is SEQ ID NO: 4, in which E. coli generally rare codons (low frequency of use) 20%) decreased from 76 of the wild gene SEQ ID NO: 2 to 43, accounting for 5.6% of the whole gene; highly rare codons (use frequency less than 10%) decreased from 46 of SEQ ID NO: 2 to 0; GC content is reduced from 68.99% ...
Embodiment 2
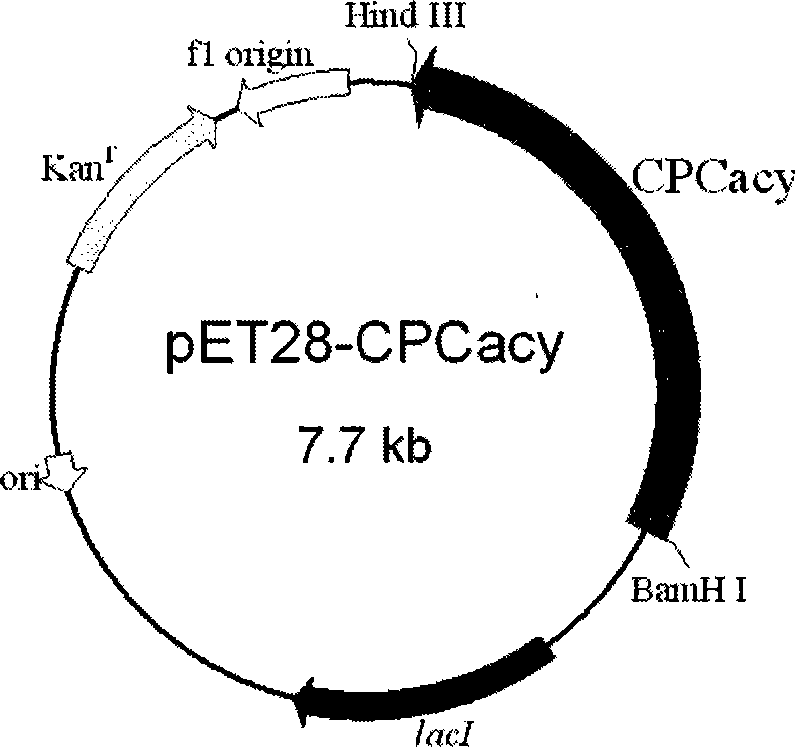
[0050] Construction of Inducible Expression Vector pET28-CPCacy and Transformant BL21(DE3) / pET28-CPCacy
[0051](1) Construction of plasmid pET28-CPCacy: the plasmid vector pUC18-acy obtained in Example 1 was digested with BamHI / HindIII double enzymes. The enzyme digestion reaction volume is 20 μL, the amount of plasmid is 4 μL, the amount of each enzyme is 1 μL, the buffer is 2 μL, and 20 μL is made up with sterile water. React overnight at 37°C to obtain the digested product CPC acylase gene. The plasmid vector pET28a (Novagen Company) was cut with the same method to obtain the linear plasmid backbone of pET28a. After purifying the digested product with a PCR product recovery kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.), the CPC acylase gene and the linearized plasmid backbone of pET28a were ligated at 4°C for 14 hours with T4 DNA ligase. The ligation reaction was transformed into competent cells of the host strain E.coli JM109, spread on LB plates (kanamycin r...
Embodiment 3
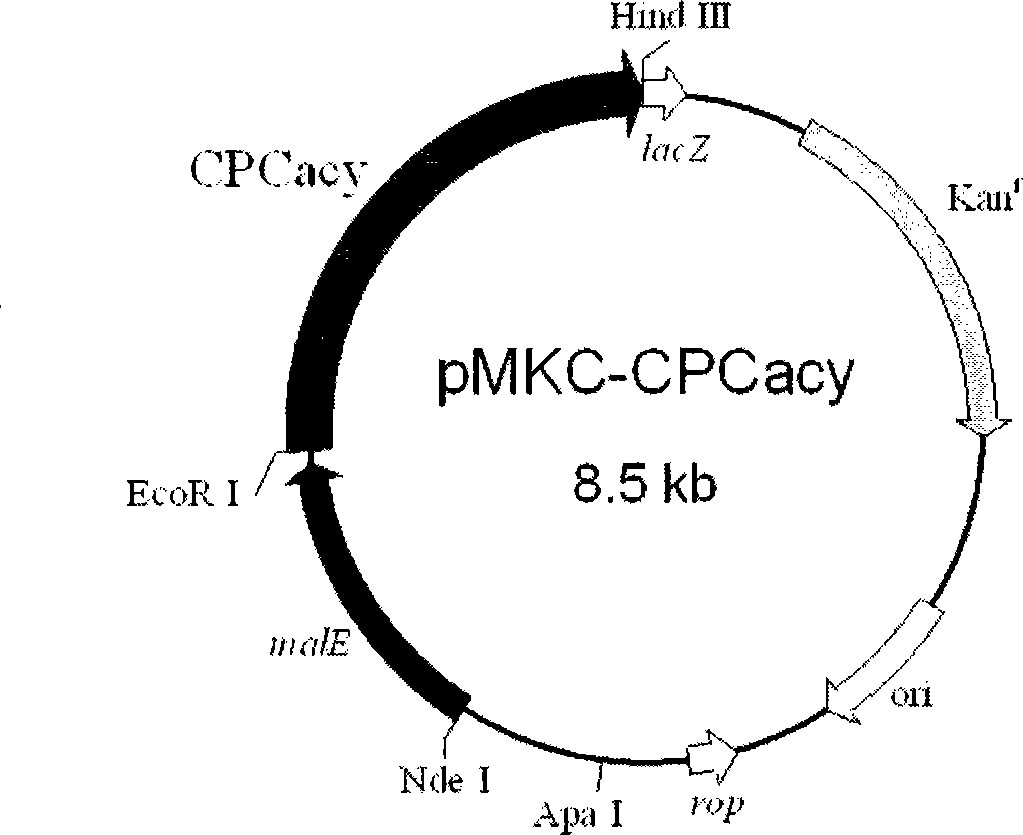
[0054] Construction of Constitutive CPC Acylase Expression Vector and Transformant
[0055] The construction method of the plasmid pMKC-CPCacy is the same as that in Example 2 (1), and the whole gene of CPC acylation enzyme is obtained by double digestion with BamHI / HindIII. At the same time, the plasmid vector pMKC-Acy (YUH.M. et al. Journal of Molecular Biocatalysis B: Enzymatic, 2006, 43, 118-123) was digested with the same method to obtain the linear plasmid backbone of pMKC. After purification with the PCR product recovery kit, the CPC acylase gene and the linearized plasmid backbone of pMKC were ligated at 4°C for 14 hours with T4 DNA ligase to construct the constitutively expressed recombinant plasmid pMKC-CPCacy (see figure 2 ). The transformation method was the same as in Example 2 (2). The electroporation transformation method was used to transform pMKC-CPCacy into Escherichia coli E. coli JM105 (Tiangen Biochemical Technology (Beijing) Co., Ltd.) to obtain the tra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com