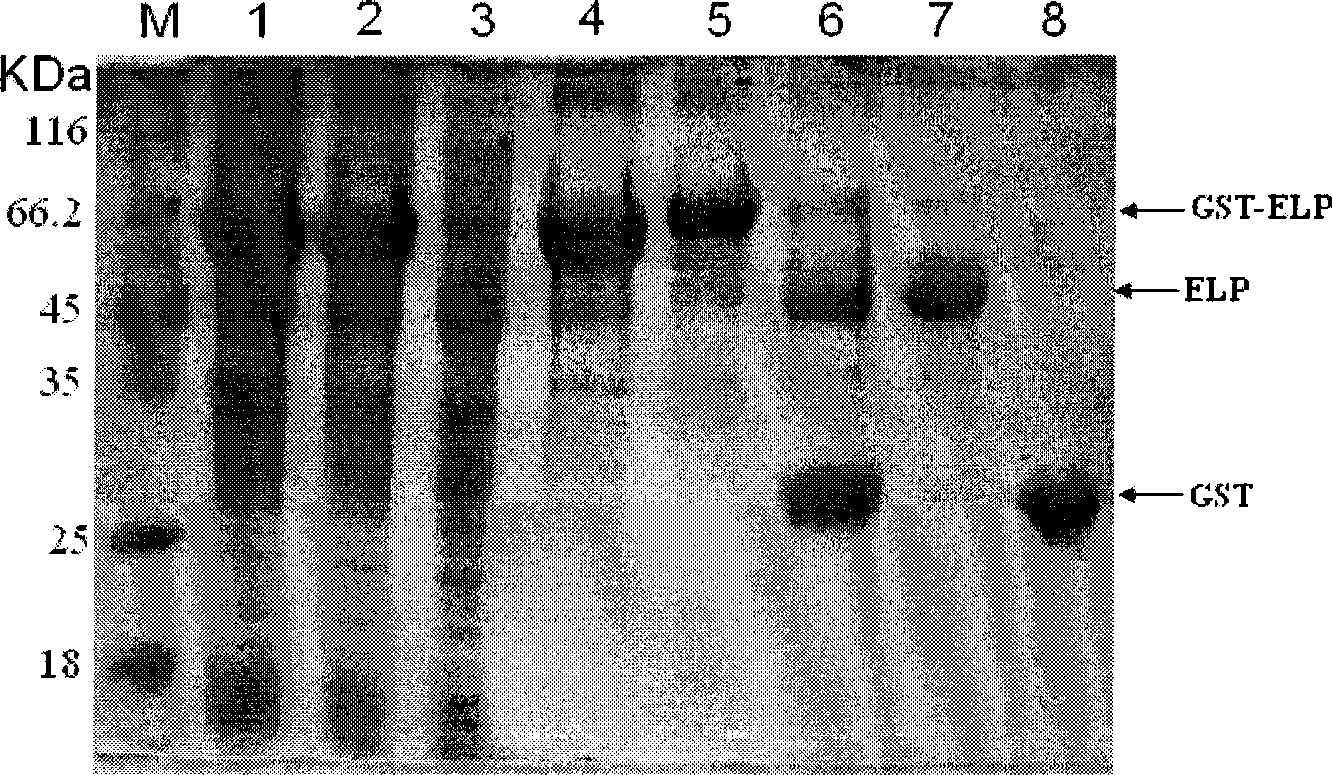High efficiency ELP fusion protease as well as preparation and application thereof
A fusion protein and protease technology, applied in the application field of cutting fusion protein, can solve the problem of high specificity of protease digestion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Construction of ELP-3C protease expression vector and transformation of host bacteria
[0032] Synthetic ELP gene (Meyer, D.E., and Chilkoti, A.1999.Purification of recombinant proteins by fusion with thermally-responsive polypeptides.NatBiotechnol 17:1112-1115.) according to the method reported by people such as Meyer and Chilkoti in 1999, its nucleoside The acid sequence is shown in SEQ ID NO.1. After being digested with EcorI at the N-terminus and HindIII at the C-terminus of the ELP gene, it was cloned into the corresponding restriction sites in the commercialized prokaryotic expression vector pET23a to form the pET23a-ELP vector. The pET23a-ELP plasmid was transformed into Escherichia coli DH5α strain, and the pET23a-ELP plasmid was extracted after culture and expression. Simultaneously, synthesize 3C protease gene according to the gene sequence (accession number: M12168) of human rhinovirus 3C protease announced on genbank in NCBI, its nucleotide seque...
Embodiment 2
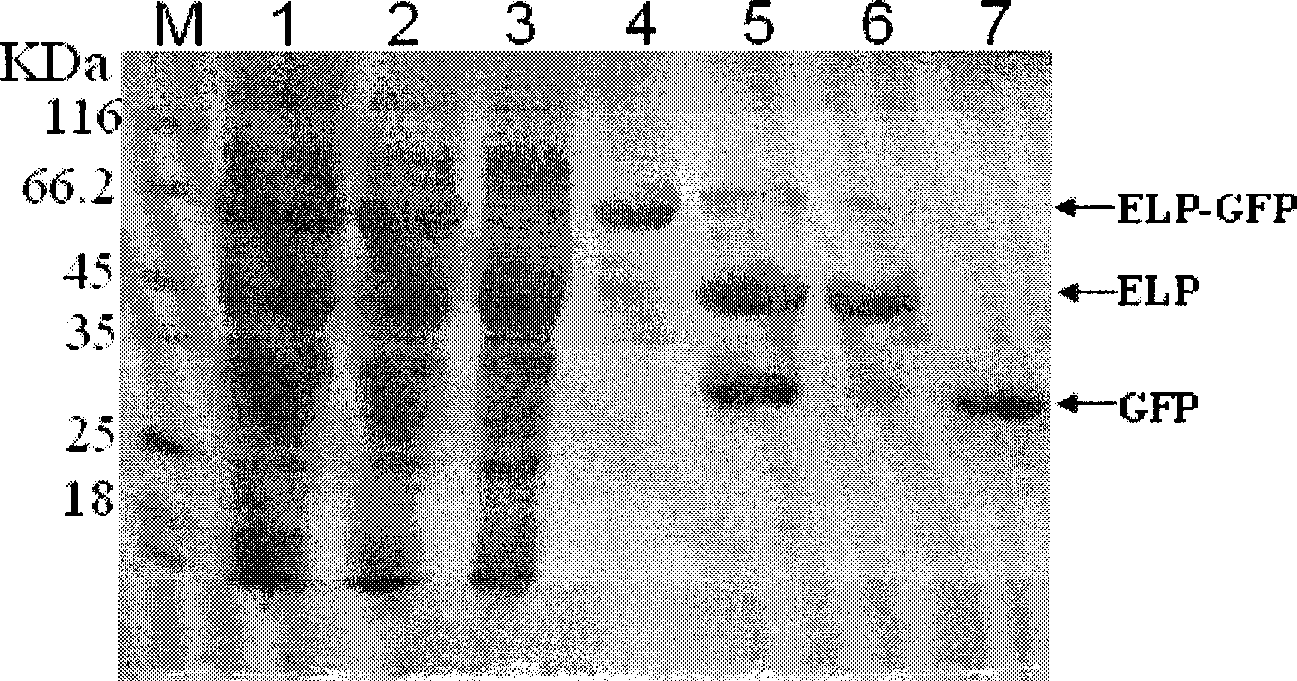
[0034] Embodiment 2: Expression and purification of recombinant ELP-3C protease
[0035] A single colony of the expression host bacterium BL21(DE3) containing the recombinant plasmid pET23a-ELP-3C was inoculated in the LB liquid enriched medium of Amp+, and cultured at 37° C. for 12 hours at 225 rpm to serve as a seed bacterium. Take the seed bacteria and inoculate them in fresh Amp+TB medium at a volume ratio of 1:100, amplify the culture with vigorous shaking at 37°C until the O.D.600 is about 1.0, then adjust the temperature to 18°C for 24 hours. 100ml of the induced bacterial solution was collected by centrifugation at 5000rpm for 5 minutes. The cells were resuspended with 8ml of ice-cold PBS, and then the cells were lysed by sonication at a power of 200W for 15 minutes. The lysed bacteria solution was centrifuged at 12000g at 4°C for 10 minutes to remove insoluble bacterial protein. Add solid NaCl to the centrifuged supernatant and dissolve it. The concentration of th...
Embodiment 3
[0036] Embodiment 3: utilize ELP-3C protease to purify GST protein
[0037] A single colony of pGEX-ELP expression host bacterium BL21(DE3) containing the recombinant plasmid was inoculated in the LB liquid enriched medium of Amp+, and cultured at 37°C and 225rpm for 12 hours as a seed bacterium. The seed bacteria were inoculated in fresh Amp+LB enriched medium at a volume ratio of 1:100, amplified and cultured with vigorous shaking at 37°C until the O.D.600 was about 1.0, then 0.5mMIPTG was added, and the expression was induced at 37°C for 4 hours. After the bacteria were harvested, the solution was lysed, and the concentration of the solution was 2M by adding solid NaCl to trigger the GST-ELP to agglutinate and centrifuge to precipitate it, and 3C digestion buffer was used for dissolution. Add ELP-3C protease at a ratio of 1:100, and digest at 5°C for 16 hours. After digestion, solid NaCl was added to make the solution concentration 2M, triggering the aggregation of ELP and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com