Primer set for amplification of CYP2C19 gene, reagent for amplification of CYP2C19 gene comprising the same, and use of the same
A CYP2C19, gene amplification technology, applied in recombinant DNA technology, fermentation and other directions, can solve the problems of low reliability of analysis results, unpractical analysis, and a lot of labor, and achieves omitting pretreatment, shortening amplification reactions, and reducing labor and labor. cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
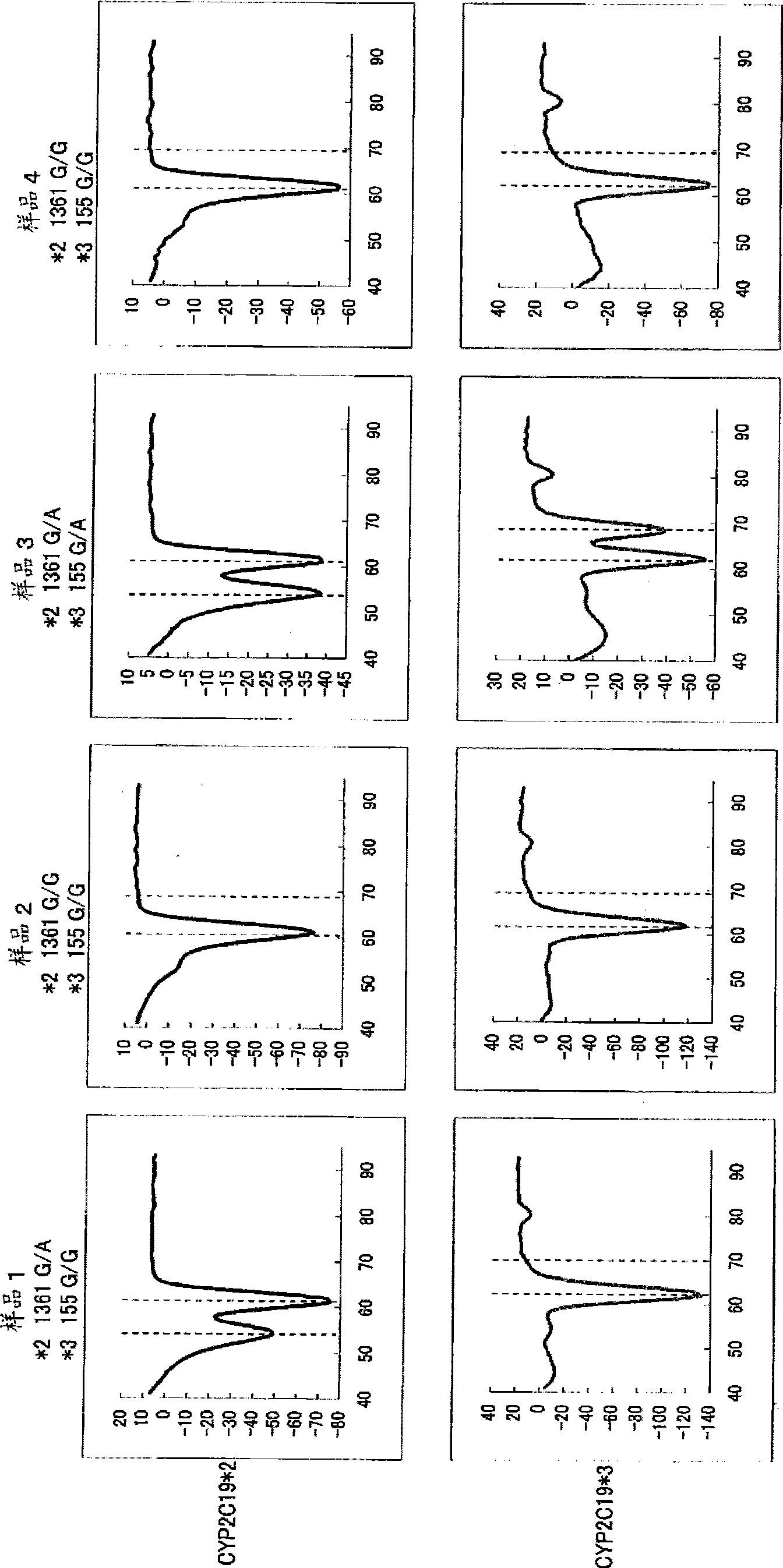
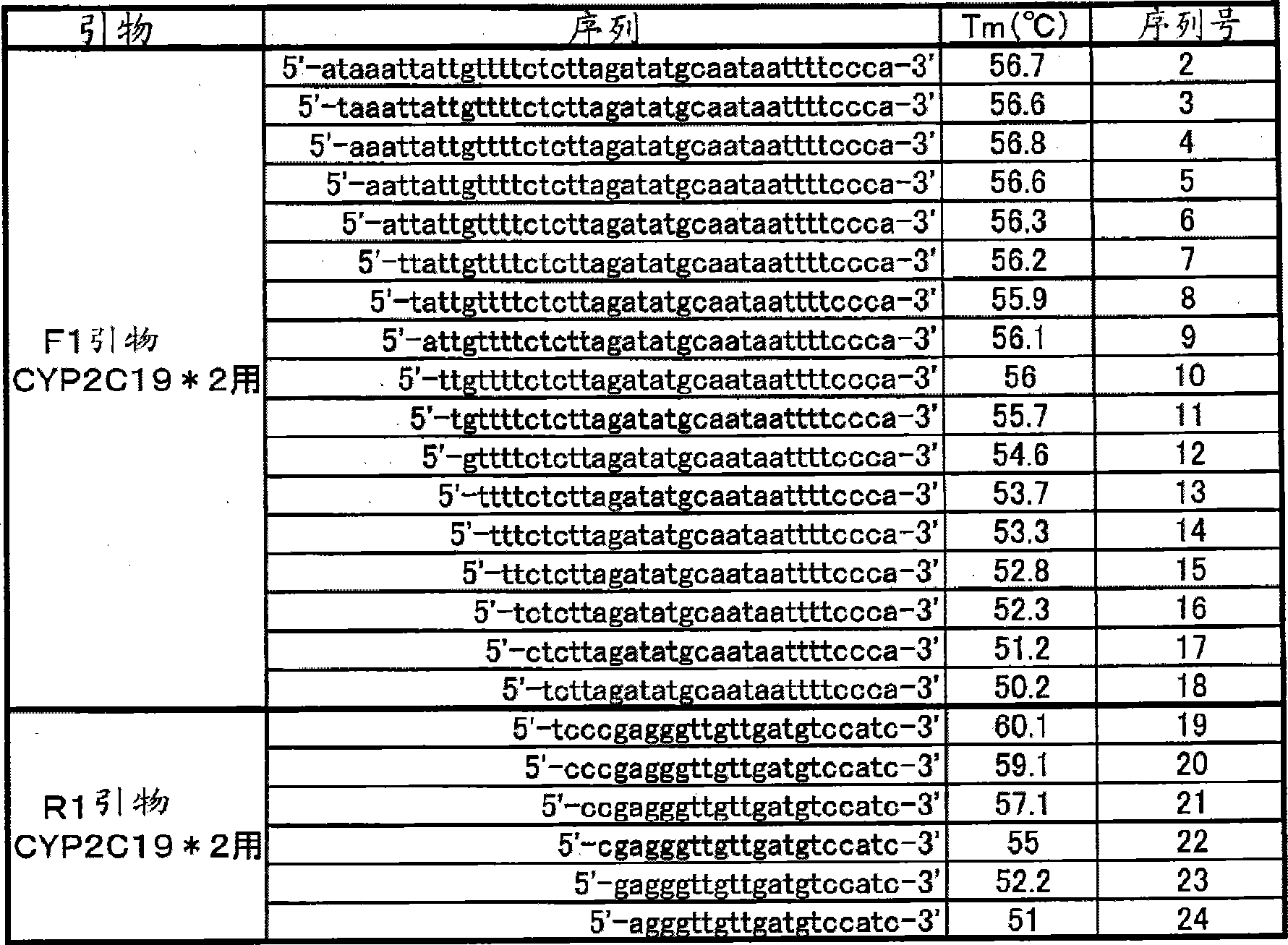
[0146] Blood was collected from 4 subjects using lithium heparin blood collection tubes (samples 1 to 4). 10 μL of the obtained blood was mixed with 90 μL of distilled water, and 10 μL of the mixed solution was mixed with 90 μL of distilled water. 10 μL of this mixed solution was added to 40 μL of a PCR reaction solution having the following composition, and PCR was performed using a thermal cycler. The conditions of PCR were as follows: after treatment at 95°C for 60 seconds, 1 cycle at 95°C for 1 second and 10 seconds at 54°C was repeated for 50 cycles, and then at 95°C for 1 second, and then Treatment at 40°C for 60 seconds. Next, the above-mentioned PCR reaction solution was heated from 40° C. to 95° C. at a rate of temperature increase of 1° C. / 3 seconds, and changes in fluorescence intensity over time were measured. The measurement wavelengths are 450-480 nm (detection of the fluorescent dye Pacific Blue), and 515-555 nm (detection of the fluorescent dye BODIPY FL). N...
Embodiment 2
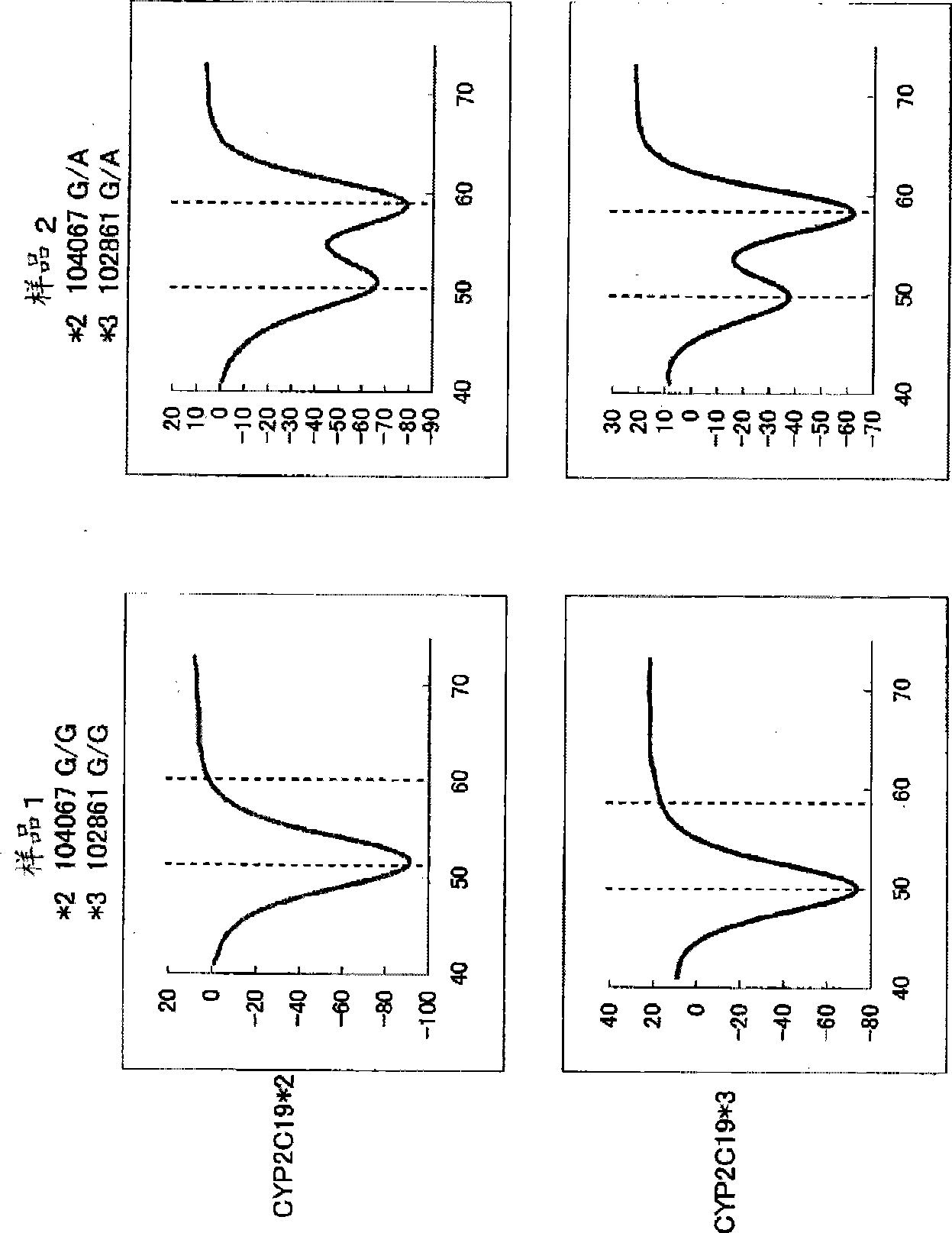
[0169] Blood was collected from two subjects using EDTA blood collection tubes (Samples 1-2). 10 μL of the obtained blood was mixed with 70 μL of the following diluent A, and 10 μL of the mixed solution was mixed with 70 μL of the following diluent B. 10 μL of this mixture was heat-treated at 95° C. for 5 minutes, then added to 46 μL of a PCR reaction solution having the following composition, and PCR was performed using a thermal cycler. The conditions of PCR are as follows: after treating at 95°C for 60 seconds, repeating 50 cycles of 1 second at 95°C and 15 seconds at 64°C, and then treating at 95°C for 1 second, Treat at 40°C for 60 seconds. Next, the above-mentioned PCR reaction solution was heated from 40° C. to 75° C. at a rate of temperature increase of 1° C. / 3 seconds, and changes in fluorescence intensity over time were measured. The measurement wavelength is 515-555nm (detection of fluorescent dye BODIPY FL) and 585-700nm (detection of fluorescent dye TAMRA).
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com