Composition for detecting O1 group vibrio cholerae, kit and detection method
A technology of Vibrio cholerae and composition, which is applied in the field of specific detection composition for O1 group Vibrio cholerae, can solve the problems of false positive results, insufficient specificity of primers, false negative results, etc., so as to reduce biological hazards and save The effect of manpower and time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
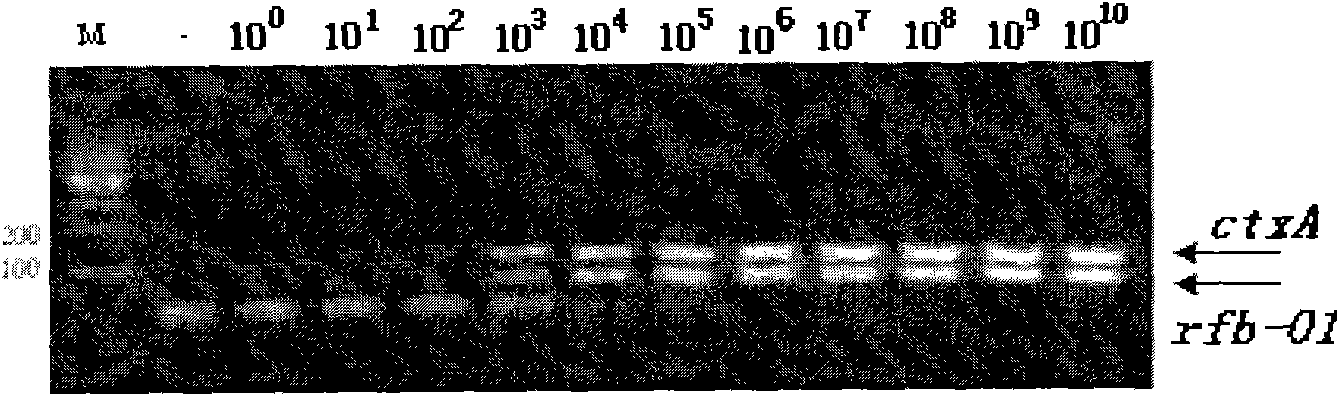
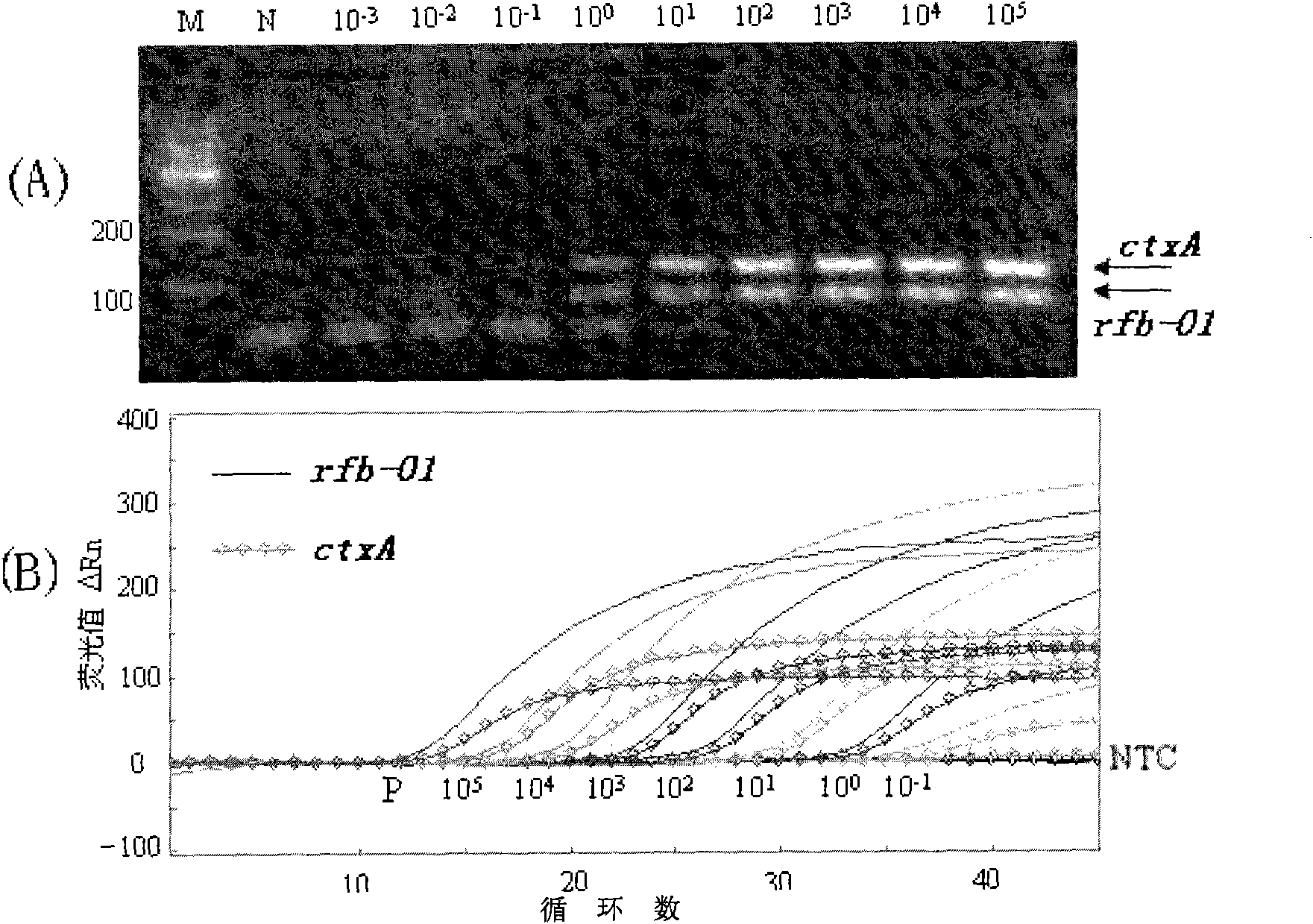
[0042] Embodiment 1 adopts the detection of common PCR method
[0043] Extraction of Bacterial Genomic DNA
[0044] The genomic DNA of each bacteria was extracted (MiniBEST kit from TAKARA Company). Use ultraviolet spectrophotometer to measure the purity and concentration of each bacterial genome DNA, O1 group Vibrio cholerae genome DNA was diluted to 1×10 with TE buffer 5 pg / μl to 1×10 -3 pg / μl gradient standard, the genome DNA of other 19 kinds of other intestinal pathogenic bacteria or common pathogenic bacteria in hospital infection was diluted to 5×10 with TE buffer 4 pg / μl, all kinds of genomic DNA were aliquoted in small quantities and stored at -20°C for future use.
[0045] Construction and preparation of plasmid standards
[0046] The target plasmid was constructed by TA cloning technique. After the molecular weight of the amplified fragment was confirmed by electrophoresis, the PCR products of rfb-O1 and ctxA specific sequences were connected to the pMD18-T vec...
Embodiment 2
[0049] Embodiment 2 adopts fluorescence real-time quantitative PCR reaction method to detect
[0050] Using the same bacterial genomic DNA and plasmid standard as in the examples, the difference is that the PCR method used is fluorescent real-time quantitative PCR, and the probes designed and synthesized above are used.
[0051] Real-time fluorescence duplex PCR reaction conditions
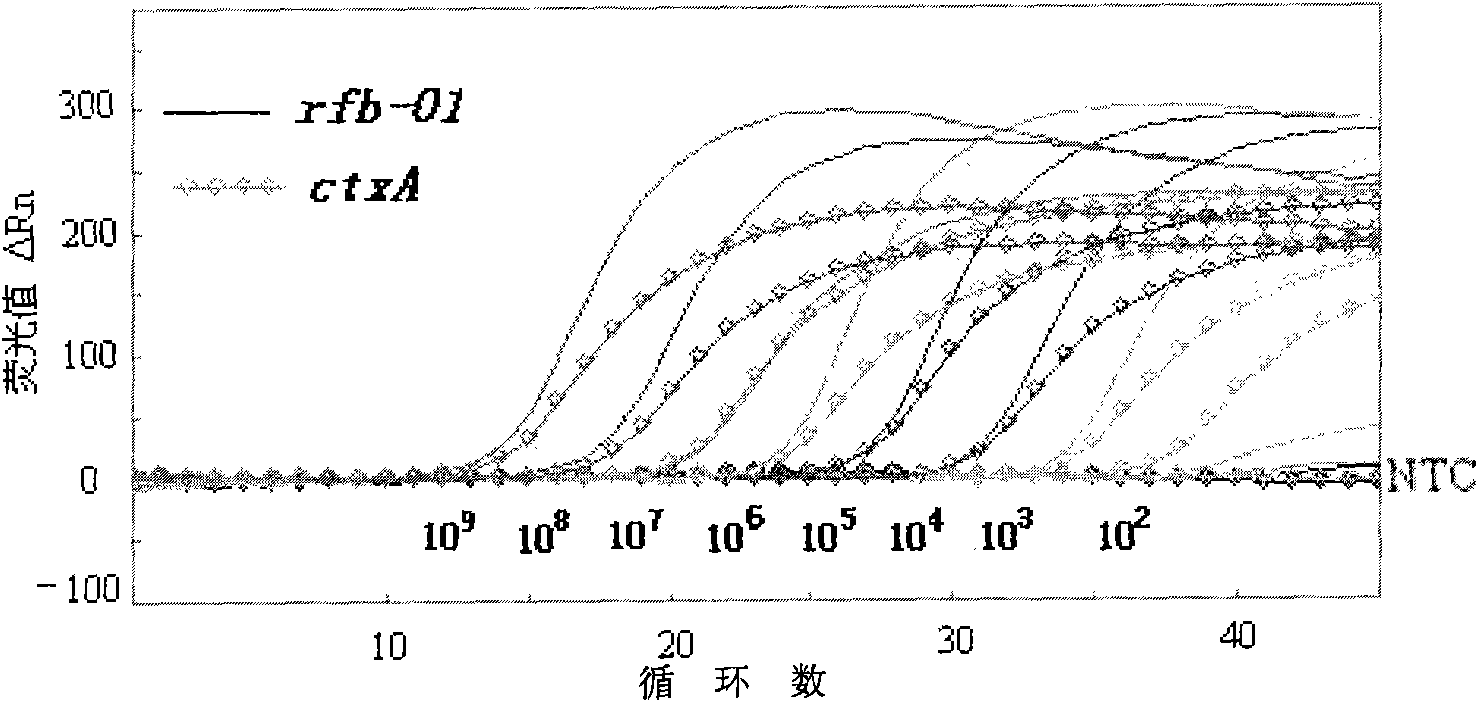
[0052] The reaction system is 25 μl, take 2.5 μl of 10×PCR buffer, 2.0 μl of each dNTP of 2.5 mmol / L, 0.5 μl of each 25 μmol / L primer, 0.5 μl of each 5 μmol / L probe, 1 U of Blend Taq plus DNA polymerase, template DNA 10ul, add sterilized water to 25μl of the final system. PCR cycle parameters: pre-denaturation at 94°C for 2min, denaturation at 94°C for 10s, extension at 60°C for 30s, a total of 45 cycles of reaction. After the amplification was completed, the data was analyzed under the same conditions to determine the Ct value of each sample.
[0053] The evaluation of the detection sensitivit...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com