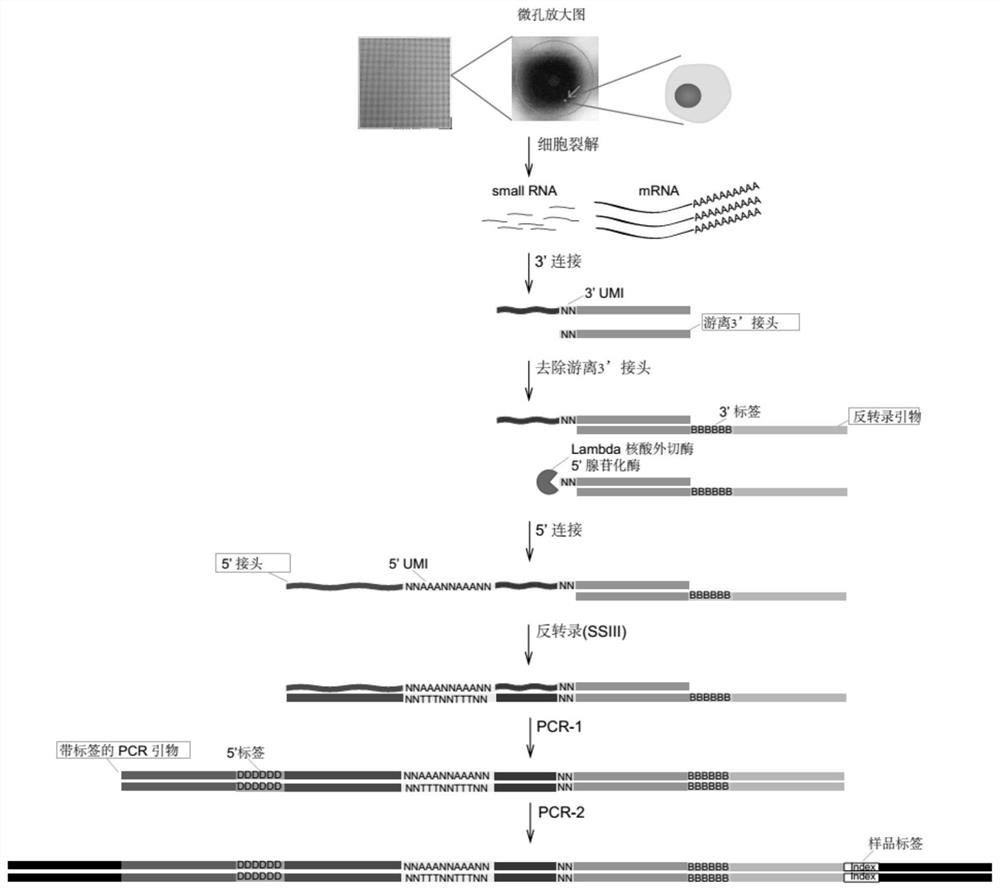
A high-throughput single-cell small RNA library construction method
A library construction, single-cell technology, applied in libraries, chemical libraries, nucleotide libraries, etc., can solve the problems of high self-ligation yield of linkers, preference for ligation reaction, and inability to achieve single-cell, so as to improve the ligation efficiency. and accuracy, no cell type specificity, improved reverse transcription efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
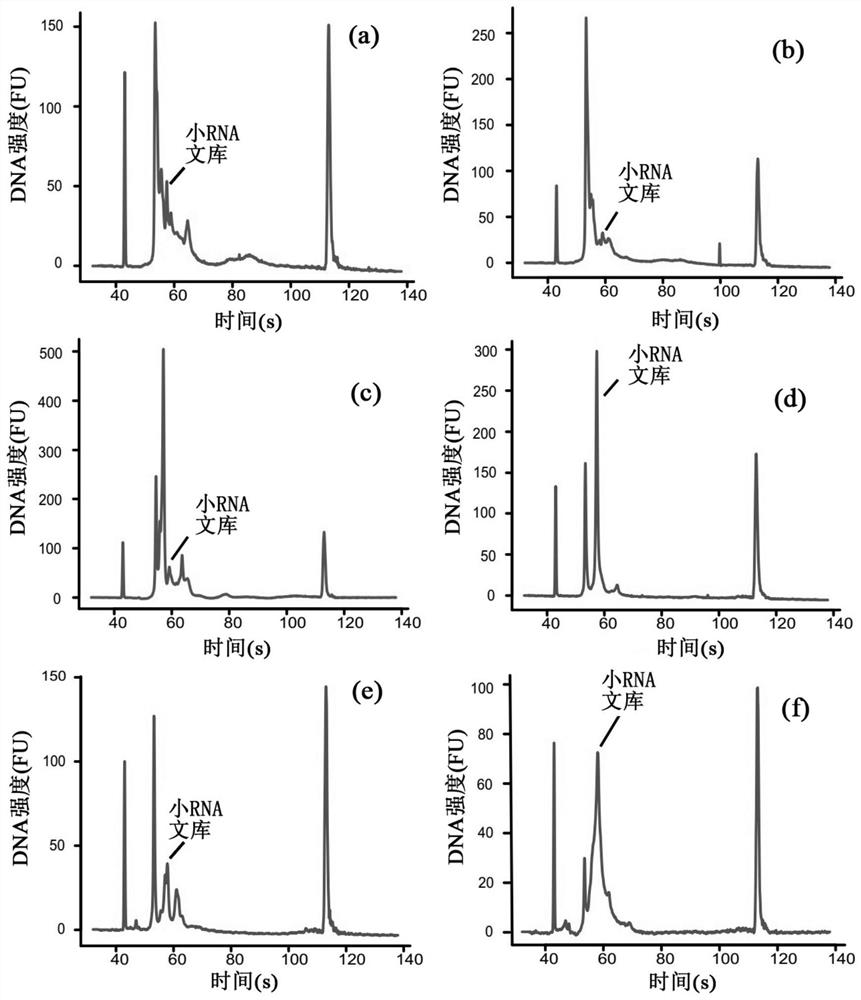
[0302] Example 1 The high-throughput single-cell small RNA library construction of A549 cells (flow chart as shown in figure 1 shown)
[0303] (1) Prepare A549 cells
[0304] 10% (v / v) fetal bovine serum and 1% penicillin-streptomycin were added to DMEM / F-12 basic medium as the culture environment of A549 cell line. The A549 cell line was cultured in a humidified incubator with an ambient temperature of 37°C and a carbon dioxide content of 5%. During the experiment, fresh cells were taken, washed twice with 1X phosphate-buffered saline (DPBS) water, and then suspended in 1X DPBS containing 0.04% bovine serum albumin. Cell suspensions were stained with 4',6-diamidino-2-phenylindole (DIPA) to mark dead cells. BD FACSAriaIII flow cytometer was used to sort and enrich living cells to obtain cell suspension.
[0305] (2) Stain and count the cells to prepare a single cell suspension
[0306] The cell suspension was stained with Hoechst-33342 and propidium iodide, and the Ready...
Embodiment 2
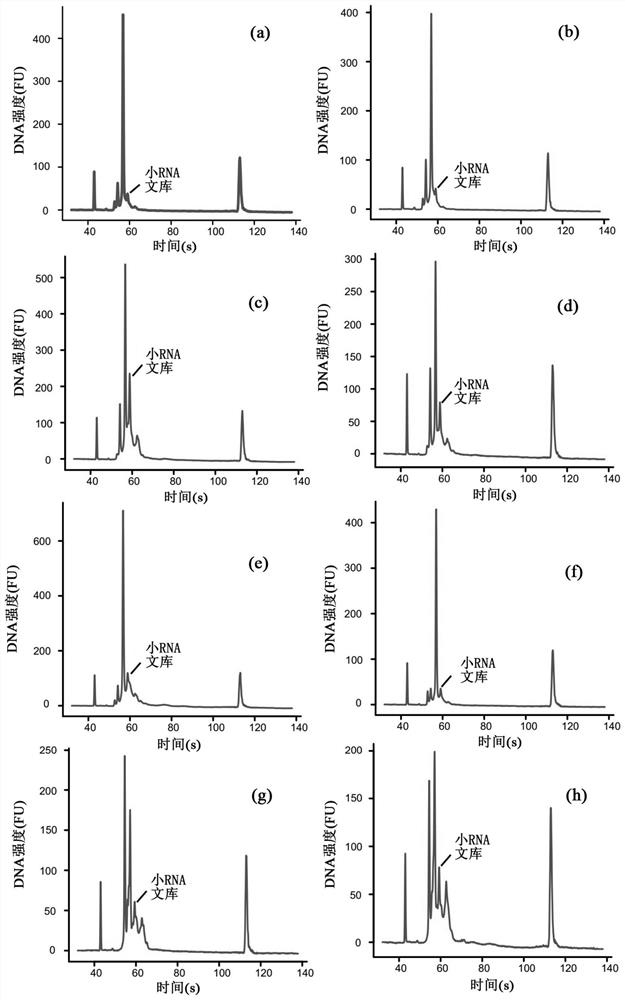
[0319] Example 2 Construction of high-throughput single-cell small RNA library of human peripheral blood mononuclear cells (PBMCs)
[0320] (1) Preparation of human peripheral blood mononuclear cells
[0321]Venous blood from healthy blood donors was collected into collection tubes containing sodium heparin anticoagulant, and peripheral blood mononuclear cells (PBMCs) were separated by Ficoll density gradient centrifugation. Heparinized blood was dissolved with an equal volume of 1x DPBS and added to a SepMateTM-15 separation tube containing an equal volume of HISTOPAQUE-1077. Centrifuge the separation tube at 1200xg for 10 minutes, quickly pour the PBMCs into a new 50ml tube, wash twice with 1xDPBS, and then suspend the cells with 1xDPBS. Cells were stained with CD45, CD3, CD19+, CD20, CD56+, CD14+, and sorted and enriched with BD FACSAriaIII flow cytometer.
[0322] A high-throughput single-cell small RNA library was constructed on human peripheral blood mononuclear cells,...
Embodiment 3
[0323] Example 3 Construction of high-throughput single-cell small RNA library of mouse B16F10 cells
[0324] (1) Culture and tumor implantation of mouse B16F10 cells
[0325] 10% (v / v) fetal bovine serum and 1% penicillin-streptomycin were added to DMEM basic medium to serve as the culture environment for the melanoma cell line B16F10. B16F10 cells were cultured in a humidified incubator with an ambient temperature of 37°C and a carbon dioxide content of 5%.
[0326] Five C57bl / 6 female mice aged 8-10 weeks were given free access to food and water under specific pathogen-free (SPF) conditions. Mice were placed in IVC cages with a 12-h light-dark cycle and fed with laboratory chow sterilized by Co60 radiation. On the day of the implantation experiment, replace the fresh medium and culture for 4 hours to collect the cells, suspend the cells in cold 1xDPBS, and the final concentration is 3x10 5 cell / ml. Inject 100ul of cell suspension into the tail vein of each animal to pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com