Recombinant fusion protein vaccine and attenuated live vector vaccine for treating and preventing helicobacter pylori (Hp) infection
A technology of Helicobacter pylori and fusion protein, which is applied in the field of recombinant fusion protein vaccine and attenuated live bacteria vector vaccine, and can solve problems such as easy generation of tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1 Obtaining HpDNA fragment cagA 160 , VacA 62 And ureB 138
[0069] 1. Primer design and synthesis
[0070] Design primers based on the nucleic acid sequence of the Hp 26695 genome (NC_000915) published by GenBank. The 5'end of the upstream primer introduces the EcoR I restriction site, the 5'end of the downstream primer introduces the Hind III restriction site, and the middle primer introduces the flexible Linker (GGGSGGGS) sequence( Picture 9 ).
[0071] The primer design is as follows:
[0072]
[0073] The primers were synthesized by Shanghai Shenggong Biotechnology Co., Ltd.
[0074] 2. PCR amplification of target DNA
[0075] 1) Preparation of template
[0076] Use "Bacterial Genomic DNA Extraction Kit" (TIANGEN, DP302) to extract the genome of Hp 26695 (purchased from American Type Culture Collection, ATCC, preserved by this unit, and can be released to the public as a verification test) genome, The operation is carried out according to the instructions.
[0077] 2...
Embodiment 2
[0081] Example 2 Construction of cagA 160 -vacA 62 -ureB 138 Fusion gene
[0082] 1. Obtain cagA by overlap extension PCR 160 -vacA 62 Fusion gene
[0083] Taking recycled cagA 160 And vacA 62 Gene fragment as template, cagA 160 P1 and vacA 62 P2 is the primer for PCR amplification, the reaction system is as follows:
[0084]
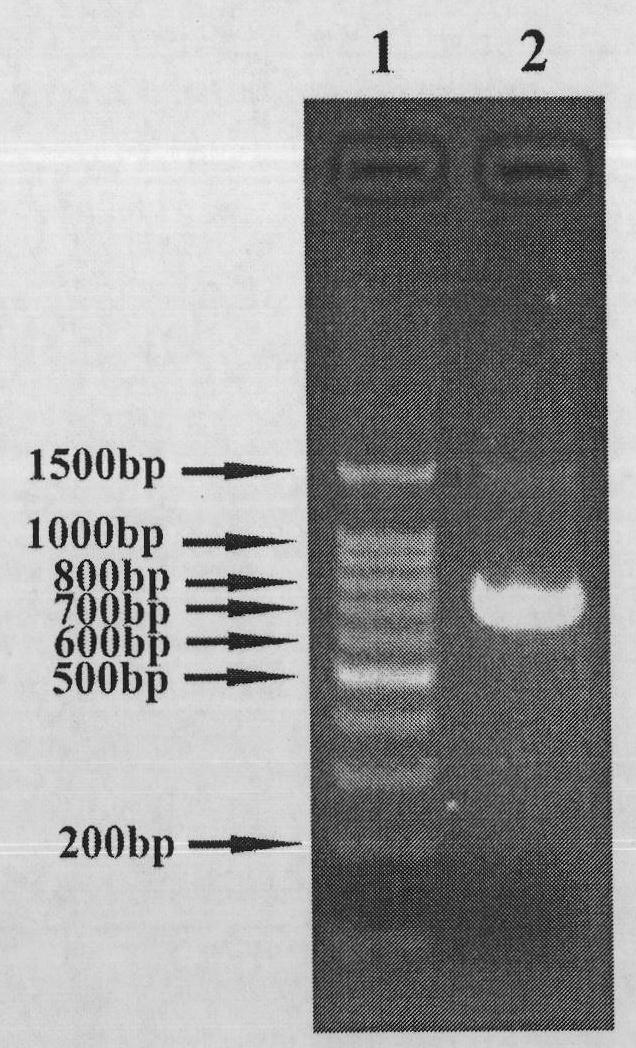
[0085] PCR reaction conditions are: 94°C pre-denaturation for 5min; 94°C denaturation for 30s, 56°C annealing for 30s, 72°C extension for 1min, 10 cycles; adding cagA 160 P1 (10μM) and vacA 62 P2 (10μM) 2μl each; denaturation at 94°C for 30s, annealing at 56°C for 30s, extension at 72°C for 2min, 30 cycles; full extension at 72°C for 10min. After the reaction is completed, take 3μl of PCR reaction product and detect it by 2.0% agarose gel electrophoresis. figure 2 The middle shows: the size of the fusion gene fragment is consistent with the prediction, indicating that the fusion gene is obtained by overlapping extension. After agarose gel electrophoresis, re...
Embodiment 3
[0090] Example 3 Recombinant plasmid pET 22b(+)-cagA 160 -vacA 62 -ureB 138 Build
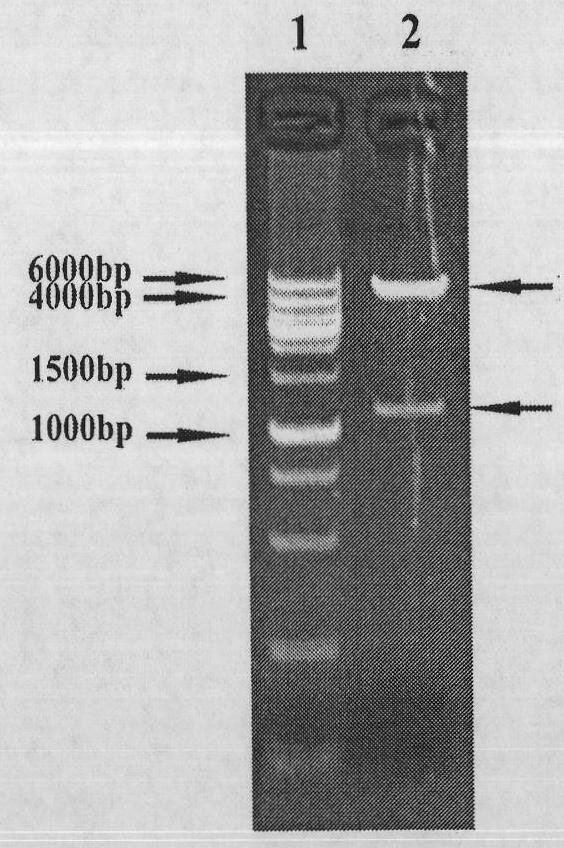
[0092] The cagA to be obtained 160 -vacA 62 -ureB 138 The fusion gene and pET 22b(+) plasmid were digested with the restriction enzymes EcoR I and Hind III of Bao Bioengineering (Dalian) Co., Ltd. respectively, and operated according to the instructions. Restriction digestion identification results such as Figure 4 Shown. Agarose gel electrophoresis, recovery of 5.5kb vector fragment and 1.1kb cagA 160 -vacA 62 -ureB 138 Fusion gene fragments.
[0093] 2. Connect
[0094] Detect the concentration of the target DNA fragment and the vector fragment by an ultraviolet spectrophotometer, and carry out the ligation reaction according to the principle that the molar ratio of the foreign fragment to the vector is generally 1:2~10 (using the ligation kit of Bao Bioengineering (Dalian) Co., Ltd. ), the connection reaction system is as follows:
[0095] cagA 160 -vacA 62 -ureB 138 E...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com