Rapid detection of BRCA (Breast Cancer) genic mutation
A gene and exon technology, applied in the fields of biotechnology and medicine, can solve problems such as pollution, low accuracy, and cumbersome detection procedures of BRCA gene mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Design, synthesis and verification of embodiment 1 primer
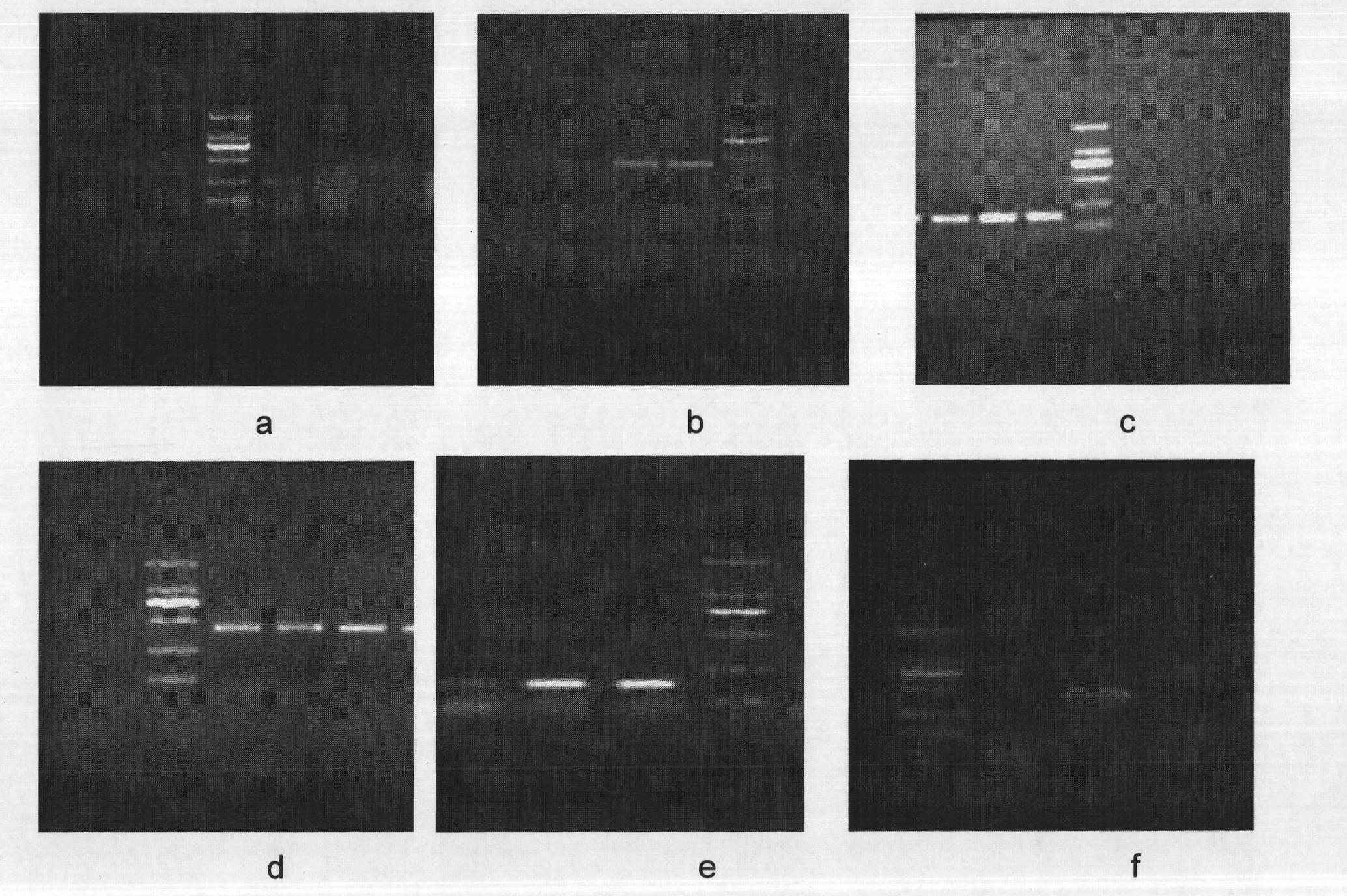
[0091] In this embodiment, Primer 5 is used to design primers. First, the size of the primer fragments is optimized to obtain the electropherograms of PCR amplification of primers with different fragment sizes. The results are as follows figure 1 and as shown in the table below. figure 1 a is the electropherogram of the primer PCR amplification of 5-9bp, figure 1 B is the electropherogram of the primer PCR amplification of 13-16bp, figure 1 c is the electropherogram of the primer PCR amplification of 18-24bp, figure 1 d is the electropherogram of the primer PCR amplification of 25-29bp, figure 1 e is the electropherogram of the primer PCR amplification of 30-37bp, figure 1 f is the electropherogram of PCR amplification with 39-45bp primers. from the table below or figure 1 It can be seen that the optimal primer is a primer with a size of 18-24bp.
[0092] Primer size (bp)
5-9
13-16 ...
Embodiment 2
[0095] The making of embodiment 2 negative or positive control DNA
[0096] Extraction of Negative Control DNA:
[0097] sampling:
[0098] 1. Take 2.5ml of peripheral blood from normal people or people with known mutations and put it in a sodium citrate (1:9) anticoagulant tube.
[0099] 2. The blood in the anticoagulant tube was centrifuged at 2000rpm for 5 minutes to separate the blood cells and washed twice with PBS.
[0100] 3. DNA extraction. The blood was extracted with the Blood Mini kit (manufactured by QIAGEN, Germany). The extraction steps were carried out according to the instructions of the kit. Other related products were treated in the same way, and the extracted DNA was stored at -20°C.
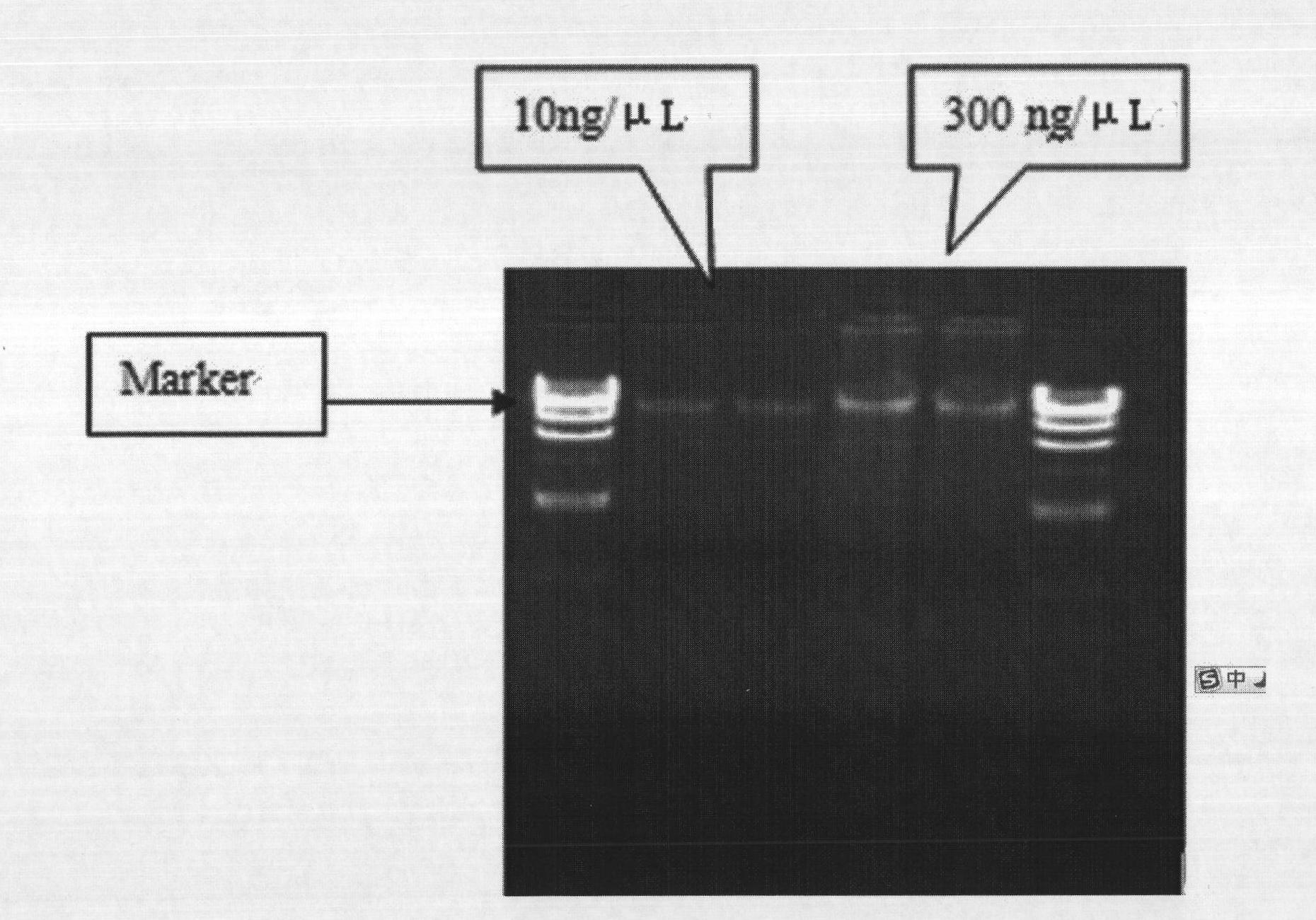
[0101] Quantification: use Nano 1000 quantifier to measure DNA concentration,
[0102] Eligibility index: 1, in line with the requirements of OD value A260 / A230: 2.0-2.2; A260 / A280: 1.8-2.0, and then quantitatively dilute the DNA to 10ng / ul
[0103] Electrophoresis identi...
Embodiment 3
[0104] Example 3 Extraction of Tumor Tissue DNA
[0105] Sample collection:
[0106] Take 50-100 mg of fresh tissue block and wash it with PBS or normal saline. The tissue used must be cut to a thickness of less than 0.5 cm, put into a cryopreservation tube containing 1.5 ml of RNAlater, and mix well. (Complete within 30 minutes) Store at room temperature for 1-2 hours, freeze overnight at 4°C, and transfer to -20°C for long-term storage the next day.
[0107] DNA extraction;
[0108] DNA was extracted using the TIANamp Genomic DNA Kit (TIANGEN BIOTECH CO., LTD) Genomic DNA Extraction Kit and following the kit instructions.
[0109] DNA purification: (if the extracted DNA, OD value A260 / A230: 2.0-2.2; A260 / A280: 1.8-2.0, is not within the standard range, this step is required)
[0110] Add an equal volume of chloroform-isoamyl alcohol (25:1), turn down the centrifuge tube for 5 minutes to mix well, and centrifuge at 13000rpm for 10 minutes to carefully suck out the supernat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com