Nucleic acid containing chimeric gene derived from hepatitis type-C virus
一种丙型肝炎病毒、嵌合基因的技术,应用在嵌合丙型肝炎病毒颗粒领域,能够解决见效性差等问题,达到高生产性、高感染性、利用价值高的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0154] Embodiment 1: Construction of TH / JFH-1 plasmid
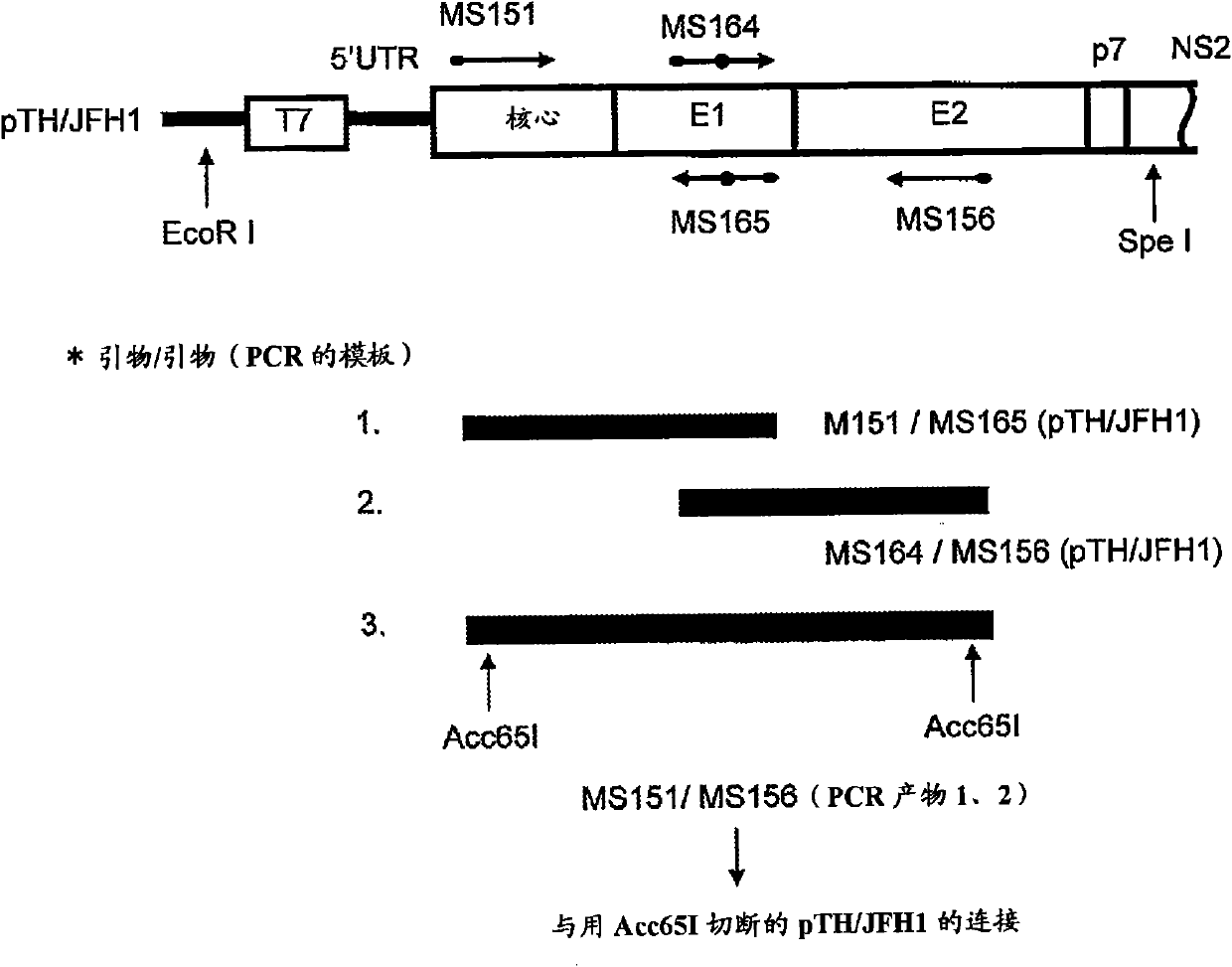
[0155] As the cDNA of HCV genomic RNA, a TH / JFH-1 chimeric cDNA was prepared, wherein the 5' UTR was the JFH-1 strain of genotype 2a (GenBank Accession No. AB047639, Kato, T. et al., Gastroenterology, 125: 1808 -1817, 2003), the N-terminal 33 amino acid residues of core protein to NS2 protein are TH strains of genotype 1b (Wakita, T. et al., J.Biol.Chem., 269:14205-14210, 1994, Moradpour, D. et al., Biochem.Biophys.Res.Commun., 246:920-924, 1998 and International Publication WO2006 / 022422), the N-terminal 34 amino acid residues ~ 3'UTR of NS2 are JFH- 1 plant.
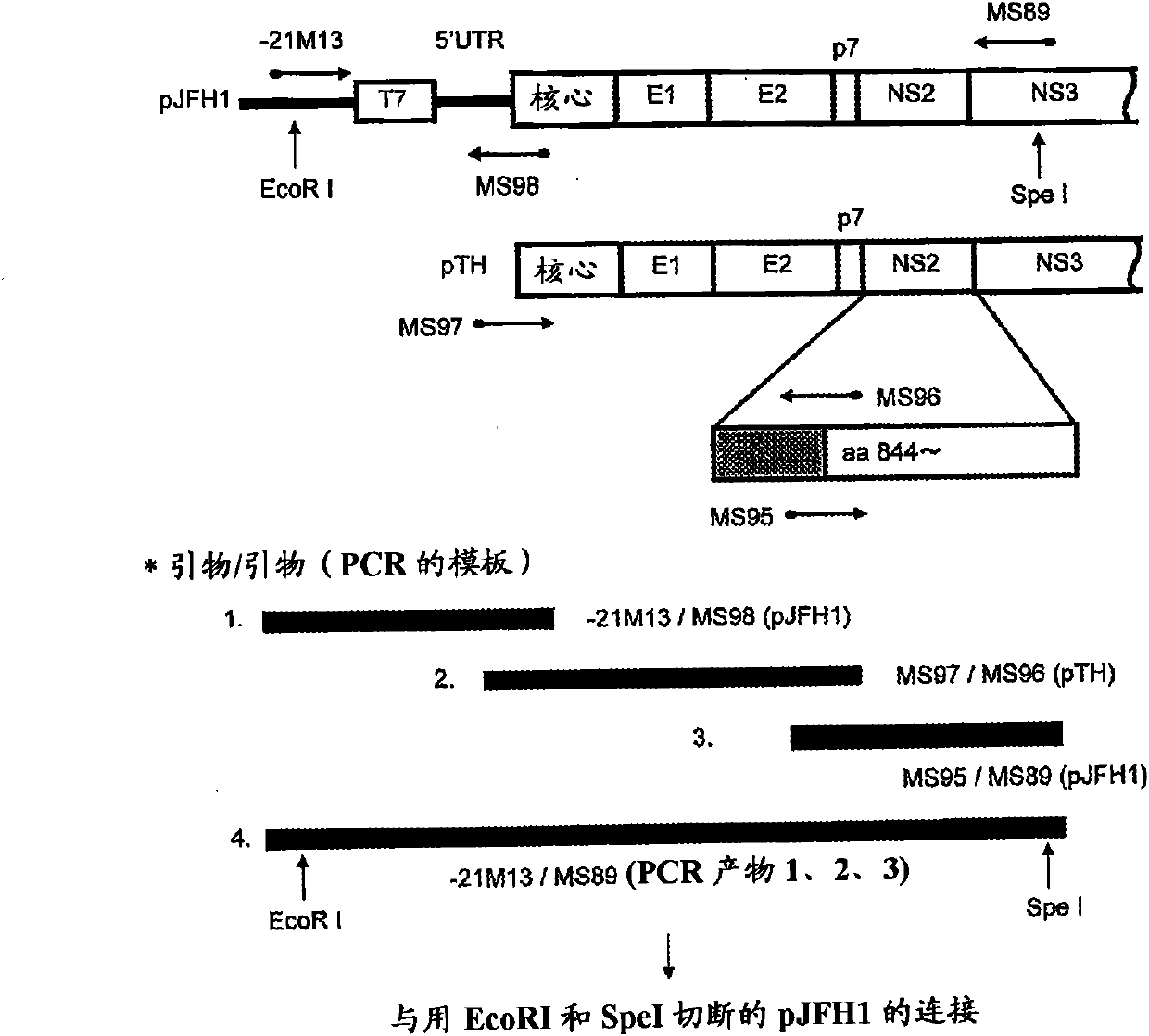
[0156] The amino acid sequence of the protein encoded by TH / JFH-1 is shown in SEQ ID NO:5. For the preparation of these plasmids, see figure 1 .
[0157] Specifically, pJFH1 (Wakita, T. et al., Nat. Med., 11:791-796, 2005, International Publication WO2004 / 104198) and pTH ( International Publication WO2006 / 022422), the pJFH1 is a plasmid DNA constructed by cl...
Embodiment 2
[0163] Example 2: In vitro RNA synthesis and its introduction into cells
[0164] pTH / JFH1 was cleaved with XbaI, extracted with phenol / chloroform, and precipitated with ethanol. Next, this XbaI cleavage fragment was treated with mung bean nuclease to remove the remaining nucleotide sequence derived from the 3' end of the XbaI recognition sequence. Then proteinase K treatment, phenol / chloroform extraction, and ethanol precipitation were performed to purify the DNA fragment. Using the cut plasmid as a template, the MEGAscriptT7 kit (Ambion) was used to react at 37°C for 3 hours to synthesize HCV RNA. After the reaction, the synthesized RNA was treated with DNaseI, extracted with acid phenol, and precipitated with ethanol to purify it.
[0165] will be 3×10 6 Huh7 cells and 10 μg HCV RNA were suspended in 400 μl Cytomix solution (120 mM KCl, 0.15 mM CaCl 2 , 10mM K 2 HPO 4 / KH 2 PO 4 , 25mM Hepes, 2mM EGTA, 5mM MgCl 2 , 20 mM ATP, 50 mM glutathione), transferred to 4 mm...
Embodiment 3
[0166] Example 3: HCV particles produced by cells introduced with TH / JFH-1 RNA
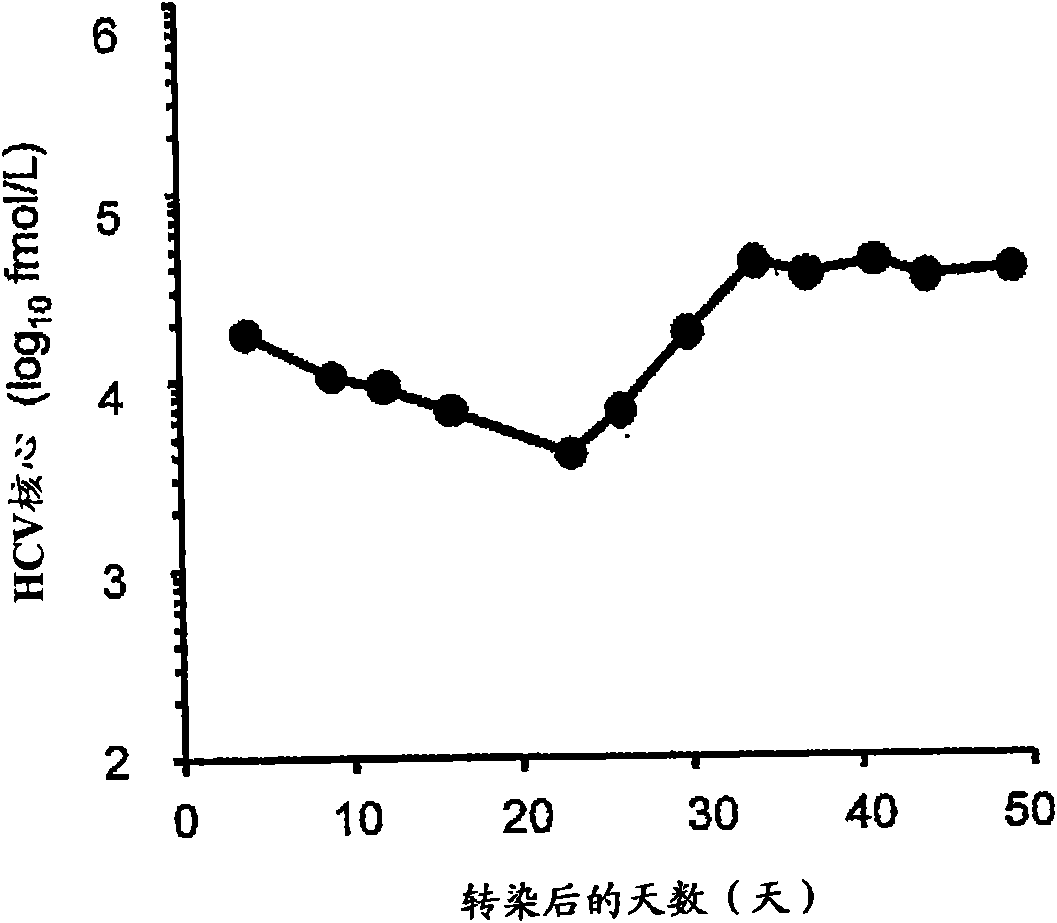
[0167] When the TH / JFH-1RNA-introduced cells were subcultured, the HCV core protein contained in the culture supernatant was quantified using the HCV antigen ELISA kit (Oiso Co., Ltd.) to confirm the production of HCV particles. As a result, the HCV core amount in the culture supernatant decreased over time until the 23rd day after the introduction, but the HCV core amount began to increase after 26 days from the introduction, and showed a constant high yield after 34 days ( figure 2 ). Therefore, it is believed that: TH / JFH-1RNA did not have high virus production ability at the beginning when it was introduced into Huh7 cells, but then the adaptive mutation necessary for virus production was introduced into the virus genome, so that TH / JFH-1RNA had high virus production ability. Virus production capacity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com