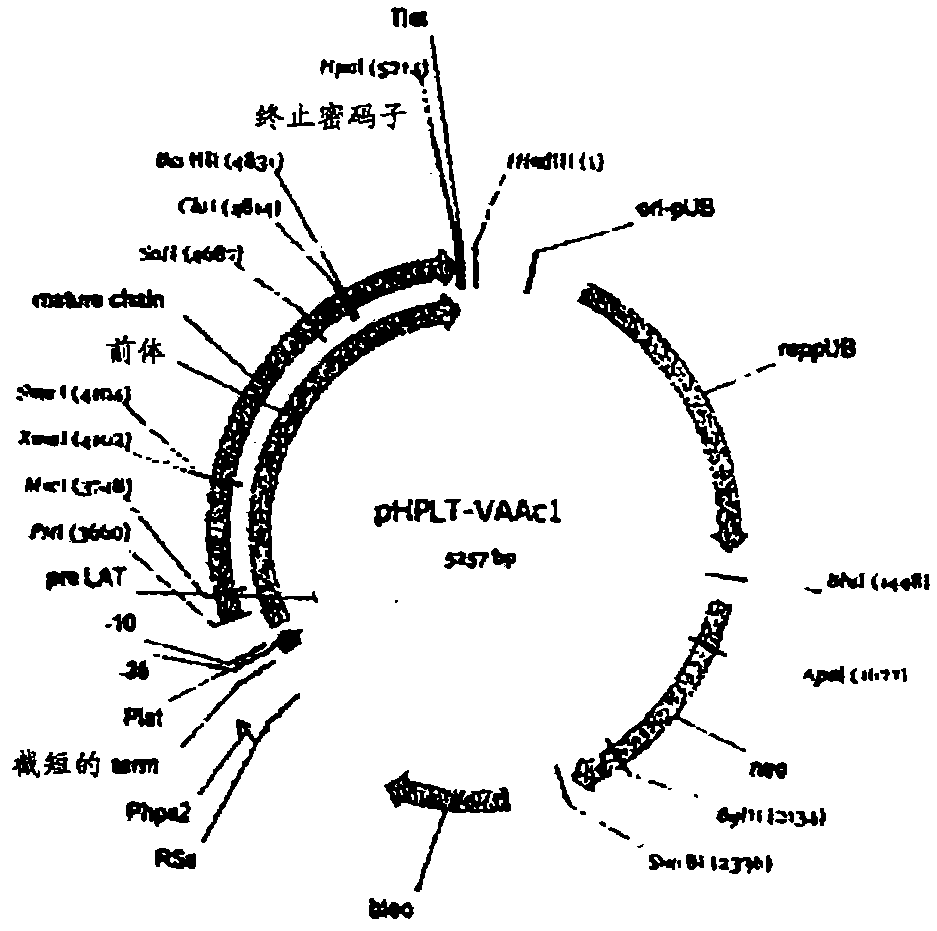
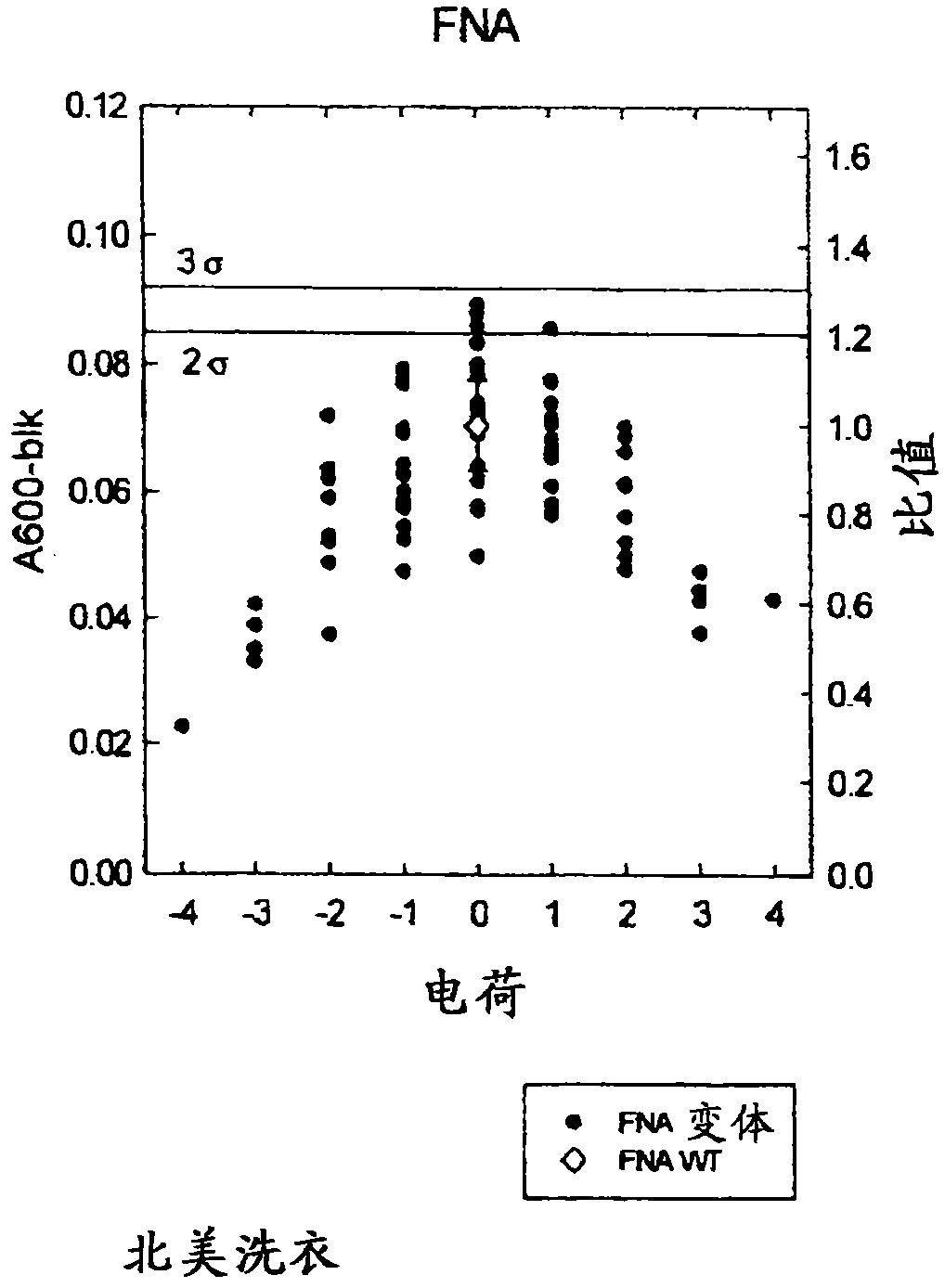
Compositions and methods comprising variant microbial proteases
A technology of protease and protease variant, applied in the field of variant protease, can solve problems such as time-consuming and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0211] Assay
[0212] In the following examples, for ease of reading, various assays are used as given below. Any deviations from the protocols provided below are indicated in the examples.
[0213] A. TCA assay for protein content determination in 96-well microtiter plates
[0214]For BPN' (e.g., reference protease) and BPN' variants, the assay was started using filtered culture supernatant from microtiter plates grown for 3-4 days at 33°C with shaking at 230 rpm and humidified ventilation . A new 96-well flat bottom microtiter plate (MTP) was used for the assay. First, place 100 µL / well of 0.25N HCl in each well. Then, 50 μL of filtered culture solution was added. Light scatter / absorbance at 405 nm (using 5 second mixing mode in the plate reader) was then measured to provide a "blank" reading. For testing, 100 [mu]L / well of 15% (w / v) trichloroacetic acid (TCA) was plated and incubated at room temperature for 5 to 30 minutes. Light scatter / absorption at 405 nm (using...
Embodiment 2
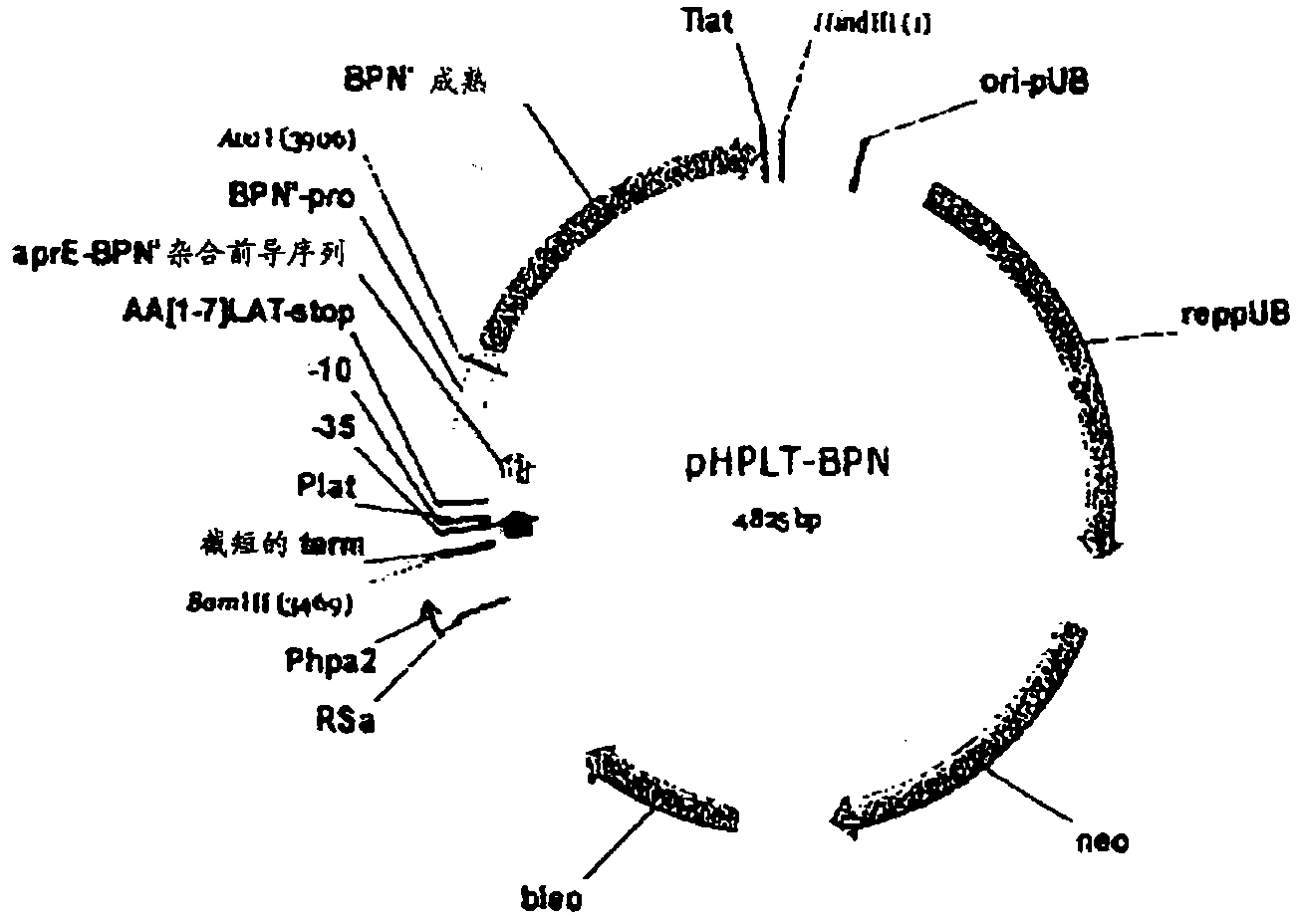
[0301] Production of BPN' protease in Bacillus subtilis
[0302] In this example, experiments performed to produce BPN' protease in Bacillus subtilis are described. Transformation of plasmid pHPLT-BPN' into Bacillus subtilis is performed as known in the art (see eg, WO 02 / 14490, incorporated herein by reference). The DNA sequence provided below (aprE-BPN' hybrid leader sequence, BPN' pre- and BPN' mature DNA sequence from Bacillus amyloliquefaciens) encodes the BPN' precursor protein:
[0303]
[0304] In the above sequences, bold indicates the DNA encoding the mature protease, standard type indicates the leader sequence (aprE-BPN' hybrid leader sequence), and underlined indicates the pre-sequence (BPN'). The amino acid sequence of the BPN' precursor protein (aprE-BPN' hybrid leader, BPN' pro and BPN' mature DNA sequences) is provided below. In this sequence, the mature BPN' protease is indicated in bold and underlined.
[0305] VRSKKLWISLLFALALIFTMAFGSTSSAQAAGKSNGEKKYIV...
Embodiment 3
[0346] Generation of BPN' site evaluation libraries and multiple mutation libraries
[0347] In this example, the construction of BPN' variants is described.
[0348] BPN' site evaluation library (SEL) construction
[0349] The pHPLT-BPN' vector containing the BPN' expression cassette was used as template DNA. This vector contains a unique BglII restriction enzyme site, which is used for SEL construction. Invitrogen Primers were synthesized (desalted, 50 nmol scale) and listed in Table 7-1 for library generation.
[0350] To construct the BPN'SEL, three PCR amplifications were performed: two mutagenesis PCRs to introduce the mutated codon of interest in the mature BPN' DNA sequence, and a third PCR fused with two mutagenesis PCRs to build in the mature BPN' sequence pHPLT-BPN' expression vector with desired mutated codons. The mutagenesis method is based on the codon-specific mutation method. In this method, generation of all possible mutations at once in a specific DNA...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Water hardness | aaaaa | aaaaa |
| Hardness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com