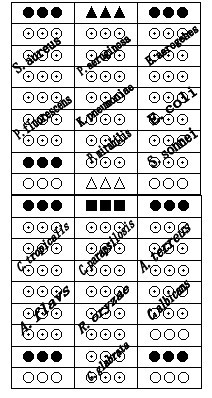
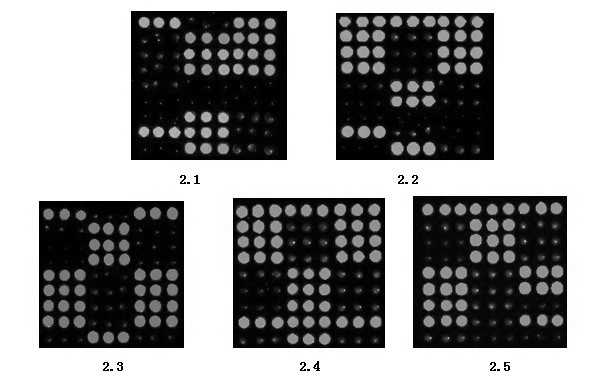
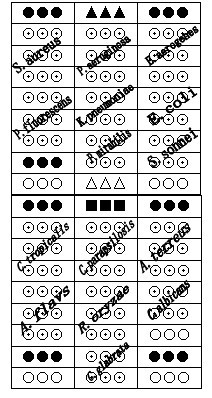
Gene chip for detecting 15 clinical common pathogenic microorganisms
A technology of pathogenic microorganisms and gene chips, which is applied in the field of gene chips for detection of common clinical pathogenic microorganisms, can solve the problems of microorganisms that cannot fully meet the purpose of accurate detection, single probes, and retention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0091] The preparation of embodiment 1 gene chip
[0092] In this example, an aldehyde-based glass slide is used as a solid phase carrier to illustrate the preparation method of the gene chip of the present invention. (1) Using aldehyde-treated glass slides as carriers;
[0093] (2) Design primers and probes:
[0094] Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Escherichia coli, Shigella, Enterobacter aerogenes, Pseudomonas fluorescens, Candida albicans, smooth Candida, Candida tropicalis, Candida parapsilosis, Aspergillus terreus, Aspergillus flavus, Rhizopus oryzae 16SrRNA gene and ITS gene sequence, through DNAMAN 5.2.2 (Lynnon Biosoft, USA) and NCBIBLAST software ( http: / / www.ncbi.nlm.nih.gov / BLAST / ) to find their conserved regions and hypervariable regions, and then use Primer Premier 5.0 (PREMIERBiosoft International, CA) to design primers in these conserved regions, Design probes in hypervariable regions. After trial an...
Embodiment 2
[0112] The detection of embodiment 2 pathogenic microorganisms
[0113] (1) Extract bacterial DNA
[0114] The DNA of pathogenic microorganisms in Table 4 was respectively extracted by CTAB / NaCl method as templates.
[0115] Table 4
[0116]
[0117] 1: ATCC: American Type Culture Collection
[0118] 2: CCTCC: China National Type Culture Collection Center
[0119] (2) target DNA amplification
[0120] The 4 pairs of primers in Tables 1 and 2 were used for PCR reaction to amplify the target DNA. The multiplex PCR reaction solution consists of 10×buffer (Biostar), 10 μmol / L of each primer, 10mmol / L dNTPs (Roche), 2U Taq DNA polymerase (Biostar), Cy3-dCTP (GE) and sterile water. The PCR reaction conditions are:
[0121] 50μl system (multiplex PCR):
[0122] Taq buffer (10×) 5μl
[0123] dNTPs (10mM) 0.8μl
[0124] srr-rl 1.5 μl
[0125] srr-dg 1.5 μl
[0126] srr-ra 1.5 μl
[0127] srr-pg 1.5μl
[0128] srr-gd 1.5 μl
[0129] srr-ak 1.5μl
[0130] Taq (2U / μl) ...
Embodiment 3
[0152] The detection of embodiment 3 pathogenic microorganisms
[0153]In this example, according to the method of Example 2, 15 strains were cultured and biochemically identified as Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Escherichia coli, Shigella, Enterobacter aerogenes, Clinical isolation of Pseudomonas fluorescens, Candida albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis, Aspergillus terreus, Aspergillus flavus and Rhizopus oryzae (Department of Laboratory, Zhongnan Hospital of Wuhan University, Wuhan, Hubei ) for the detection of pathogenic microorganisms. The results showed that the detection results were completely consistent with the results of culture and biochemical identification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com