Fluorescent quantitative PCR kit for detecting alpha-globin gene deletion
A fluorescent quantitative and globin technology, applied in the field of biochemistry, can solve the problems of operational feasibility, unsatisfactory detection throughput, false positives, false negatives, etc., and achieve good sensitivity, good specificity, repeatability, and good sensitivity and the effect of specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
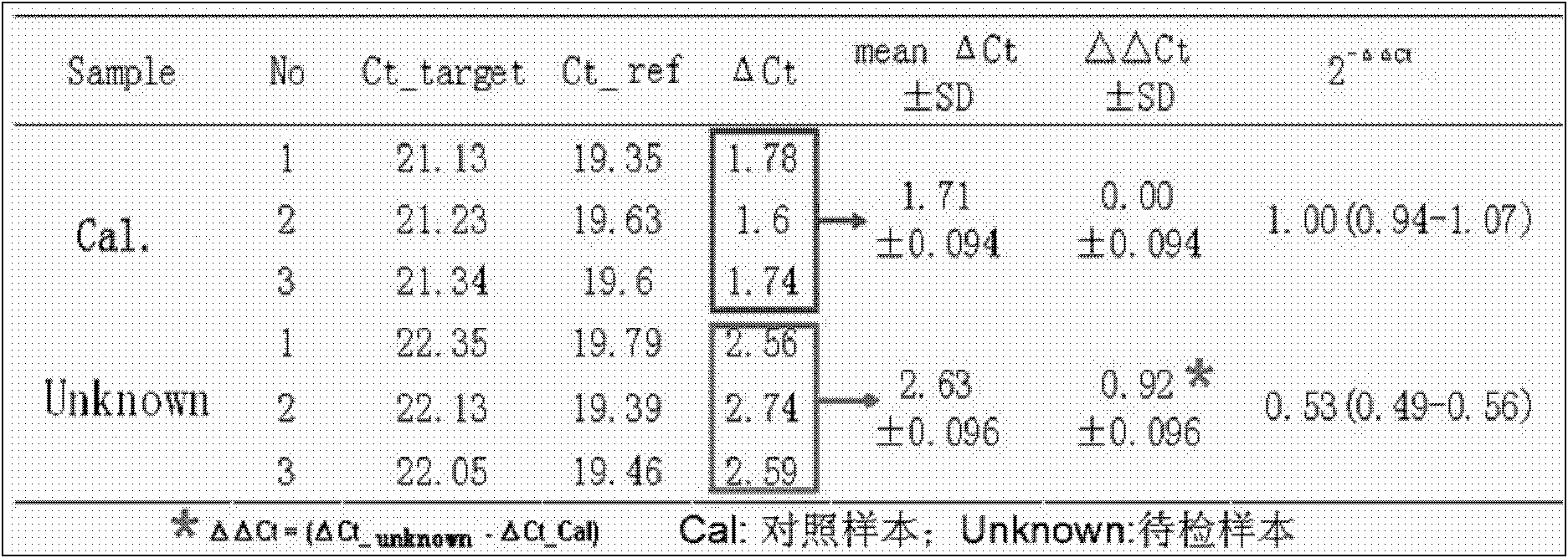
[0054] Example 1: Small sample evaluation of the sensitivity and accuracy of the kit
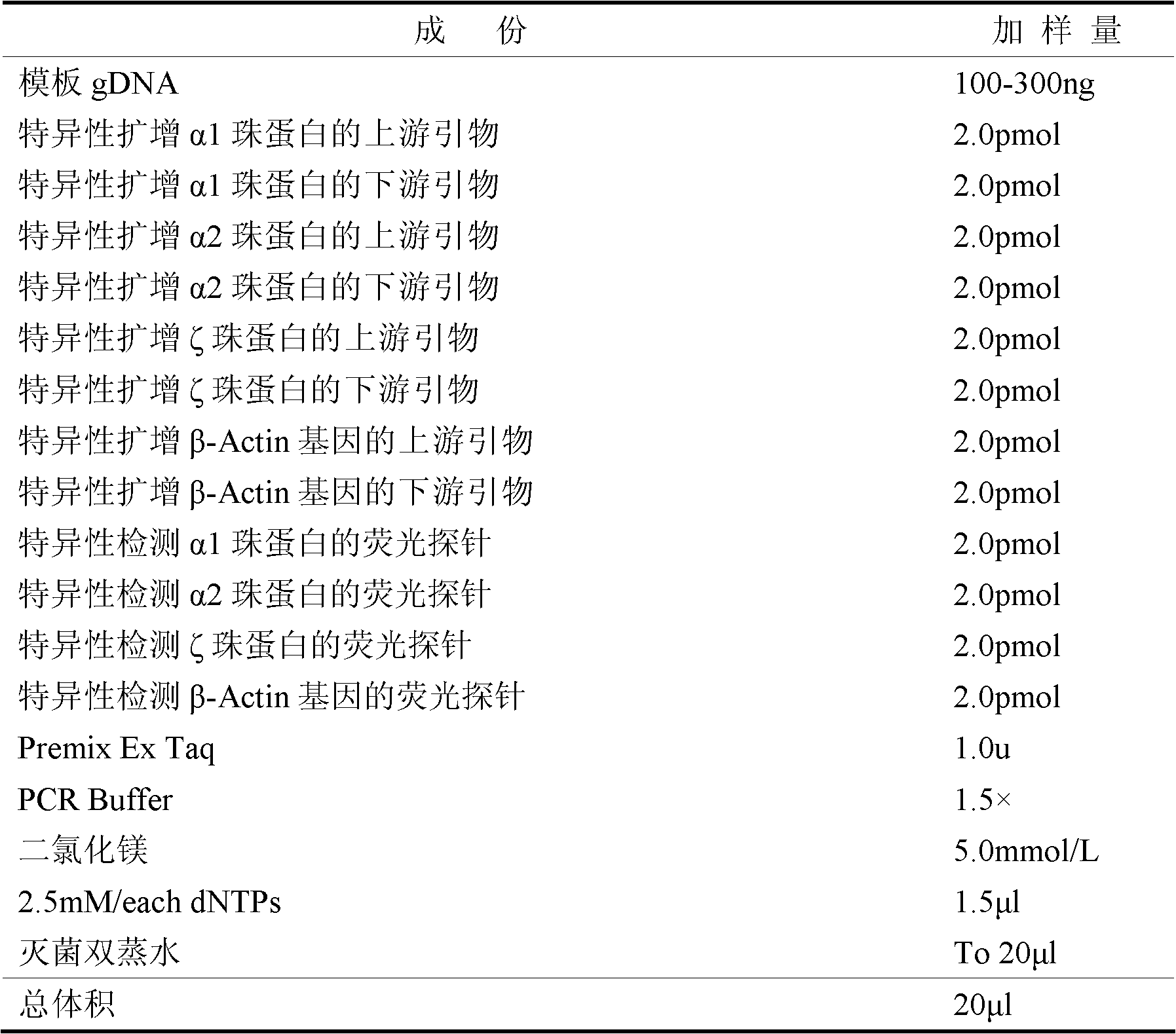
[0055] 1. The composition of the kit:
[0056] (1) The upstream primers for specific amplification of α1 globin are:
[0057] 5'-CGTCGTGTACTTGTGTGATGGTTAGA-3' (SEQ NO.1),
[0058] The downstream primers for specific amplification of α1 globin are:
[0059] 5'-CGTCGTAAACAGGTAAACAAAGCAATAG-3' (SEQ NO.2),
[0060] (2) The upstream primer sequence for specific amplification of α2 globin is:
[0061] 5'-CGTCGTGTACAACTTCCTATTCTCAGTG-3' (SEQ NO.3),
[0062] The downstream primers for specific amplification of α2 globin are:
[0063] 5'-CGTCGGGAAGACTTGCTAGGTAAATACT-3' (SEQ NO.4),
[0064] (3) The upstream primers for specific amplification of ζ globin are:
[0065] 5'-CCGTCCAGGAGGCTGCTTACT-3' (SEQ NO.5),
[0066] Downstream primers that specifically amplify zeta globin are listed as:
[0067] 5'-GGTGCTCCTTATTCATTTCAGAATCAC-3' (SEQ NO.6),
[0068] (4) The upstream primers for specifically a...
Embodiment 2
[0103] Embodiment 2: Sensitivity and specificity evaluation of the kit of the present invention
[0104] 1. Specimen collection:
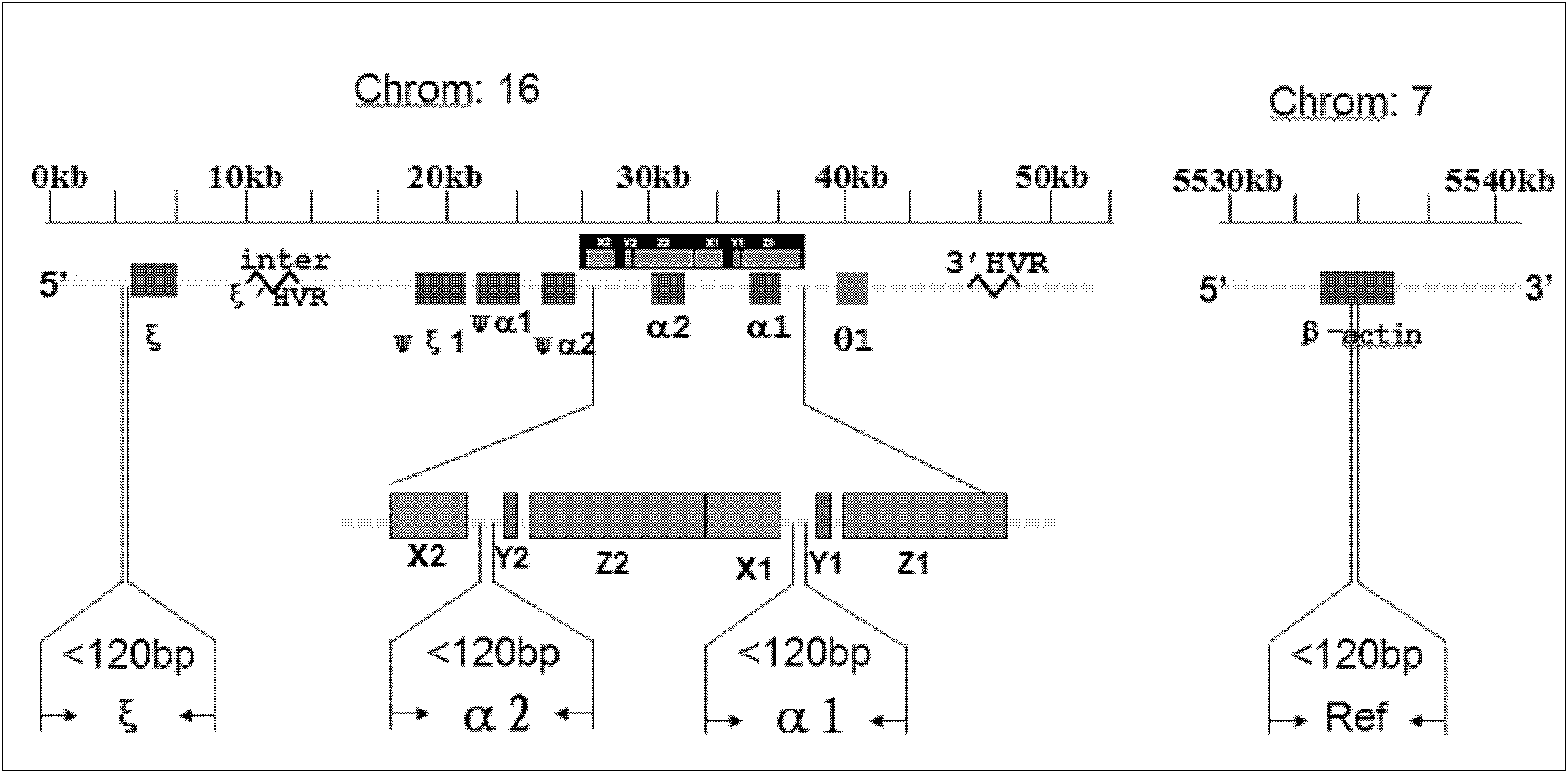
[0105] From the sample bank of our laboratory, 218 gDNA samples of known genotypes detected by gap-PCR were collected. This batch of samples contained αα / αα, -α 3.7 / αα, -α 4.2 / αα, -- SEA / αα, -- SEA / -α 4.2 、-- SEA / -- SEA 、- SEA / -α 3.7 , -α 3.7 / -α 3.7 , -α 4.2 / -α 4.2 , -α THAI For the 10 genotypes / αα, dilute the gDNA samples to 100-300ng / μl with sterilized double distilled water for later use. The gap-PCR detection system used is a detection system that has been verified by Southern Blot, and the relative copy numbers of ζ, α1, and α2 genes are accurately deduced according to the type of deletion.
[0106] 2. Specimen testing:
[0107] (1) Kit composition:
[0108] With embodiment 1.
[0109] (2) Sample detection: the above-mentioned gDNA specimens to be tested, 2 normal control gDNA samples with 2 copies of ζ, α1 and α2 glo...
Embodiment 3
[0118] Example 3: Small-scale crowd screening detection evaluation
[0119] 1. Specimen collection:
[0120] 104 peripheral blood samples were randomly collected from the Biochemical Genetics Department of the Prenatal Diagnosis Center of Nanfang Hospital, and routine blood analysis was performed on the 104 peripheral blood samples with F-820 blood analyzer to obtain phenotype data. Then use Tiangen column-type peripheral blood genomic DNA column extraction reagent (Beijing Tiangen Biotechnology Company) to extract gDNA, and dilute the gDNA sample to 100-300 ng / μl with sterilized double-distilled water for use.
[0121] 2. Specimen testing:
[0122] (1) Kit composition:
[0123] Same as Example 1
[0124] (2) Sample detection: The above 104 gDNA samples to be tested were double-blind, and the deletion α-thalassaemia gap-PCR method and this kit were used for parallel detection. The gap-PCR method used is the detection system and detection conditions that have been verified ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com