Method for simply, conveniently and quickly extracting trace total deoxyribonucleic acid (DNA) of single roes and fries
A technology for fish eggs and larvae, which is applied in the field of DNA extraction, can solve the problems of large influence on elution efficiency, large final volume of extraction, and low DNA concentration, and achieves the effect of avoiding freezing equipment, ensuring efficiency and wide application range.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
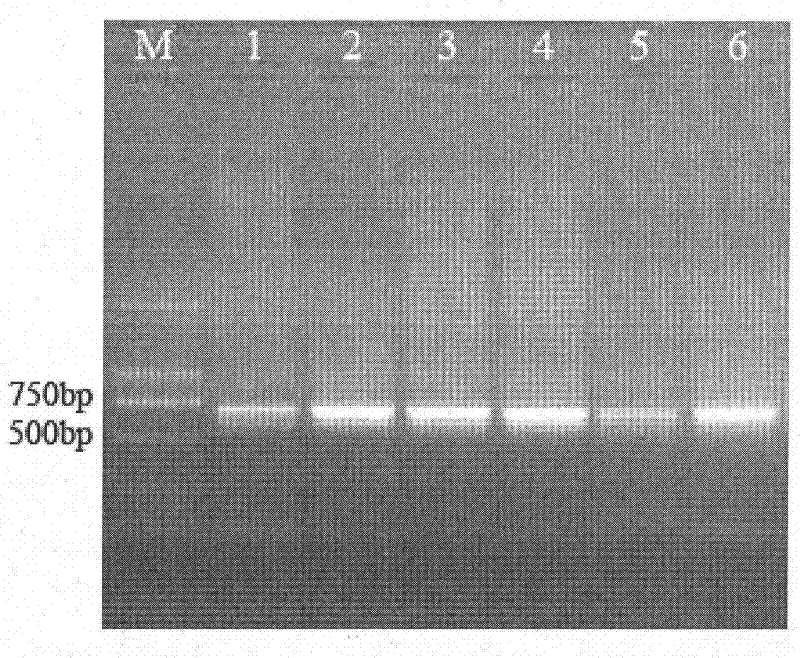
Image
Examples
Embodiment 1
[0025] 1) Sample fixation:
[0026] Collect 0.5mL of fertilized eggs of turbot, observe with a dissecting microscope that the fertilized eggs have developed to the 2-cell stage, filter out the seawater and add 1mL of absolute ethanol. After 24 hours, the anhydrous ethanol was replaced once. Store at room temperature;
[0027] 2) Removal of fixative in the sample:
[0028] Take 1 fixed fish roe obtained in step 1), add 0.9% NaCl solution in normal saline to wash, centrifuge at 1500g for 30 seconds, discard excess normal saline, dry the water with filter paper, repeat the above steps Operate 2 times;
[0029] 3) Sample digestion:
[0030] Put the washed fish eggs into a 1.5 mL centrifuge tube, add 100 μL DNA extraction buffer and 10 μL cell lysis buffer. Then use clean dissecting scissors to cut the egg membrane, add 3 μL 10 mg / mL proteinase K and 3 μL 10 mg / mL RNase. Shake and mix and incubate at 55°C, invert and mix once every 10 minutes, digest for 20 minutes until the ...
Embodiment 2
[0042] 1) Sample fixation:
[0043] Collect 1 mL of fertilized eggs of flounder, observe with a dissecting microscope that the fertilized eggs have developed to the embryo body stage, filter off the seawater and add 5 mL of absolute ethanol. After 24 hours, the anhydrous ethanol was replaced once. Store at -20°C;
[0044] 2) Removal of fixative in the sample:
[0045] Take 1 fixed fish roe obtained in step 1), add a mass-volume ratio of 0.9% NaCl solution and wash with normal saline; centrifuge at a speed of 1000g for 60 seconds, absorb and discard excess normal saline, dry the water with filter paper, repeat 2 Second-rate.
[0046] 3) Sample digestion:
[0047]Put the washed fish eggs into a 1.5 mL centrifuge tube, add 200 μL DNA extraction buffer and 20 μL cell lysis buffer. Then use clean dissecting scissors to cut the egg membrane. Add 2 µL of 20 mg / mL proteinase K and 2 µL of 20 mg / mL RNase. Shake and mix and incubate at 60°C, invert and mix once every 15 minutes, ...
Embodiment 3
[0059] 1) Sample fixation:
[0060] Collect 10 newly hatched flounder larvae, filter the seawater and add 2 mL of absolute ethanol. After 24 hours, the anhydrous ethanol was replaced once. Store at room temperature;
[0061] 2) Removal of fixative in the sample:
[0062] Take 1 fixed newly hatched larvae obtained in step 1), add a mass-volume ratio of 0.9% NaCl solution, and wash with saline; centrifuge at 2000g for 20 seconds, dry the water with filter paper, and repeat the above operation 3 times.
[0063] 3) Sample digestion:
[0064] Put the washed larvae into a 1.5 mL centrifuge tube, add 300 μL DNA extraction buffer and 30 μL cell lysis buffer. Then cut the larvae into pieces with clean dissecting scissors. Add 5 µL of 20 mg / mL proteinase K and 5 µL of 20 mg / mL RNase. Shake and mix and incubate at 55°C, invert and mix once every 20 minutes, digest for 1 hour until the solution is clear; the DNA extraction buffer is pH=8.0 with a final concentration of 0.01mol / L, Tr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com