Primer for amplifying specific regions of common pathogenic type Legionella gyrB genes and application thereof
A technology of Legionella and pathogenic type, applied in the field of Legionella Mikdide, can solve the problems of high price, time-consuming and laborious, unfavorable rapid diagnosis and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Example 1 : Genome Extraction
[0080] In BCYE plate medium at 37°C, 5% CO 2 Culture Legionella Mickel-Daider, Legionella bozemannii and Legionella longbeach respectively overnight, scrape the colonies with 50mM Tris-HCl (pH8.0), and collect the bacteria by centrifugation. The cells were resuspended with 500ul 50mM Tris-HCl (pH8.0) and 10ul 0.4M EDTA, incubated at 37°C for 20 minutes, and then 10ul 10mg / ml lysozyme was added to continue the incubation for 20 minutes. Then add 3ul 20mg / ml proteinase K, 15ul 10% SDS, incubate at 50°C for 2 hours, then add 3ul 10mg / ml RNase, and incubate at 65°C for 30 minutes. Add an equal volume of phenol to extract the mixture, take the supernatant, and then extract twice with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), take the supernatant, and then extract with an equal volume of ether. Extract to remove residual phenol. The supernatant was used to precipitate DNA with 2 times volume of ethanol, and the DNA wa...
Embodiment 2
[0081] Example 2 : Amplification of the gyrB gene from Legionella meek-daid, Legionella bozemannii, Legionella longbeach by PCR
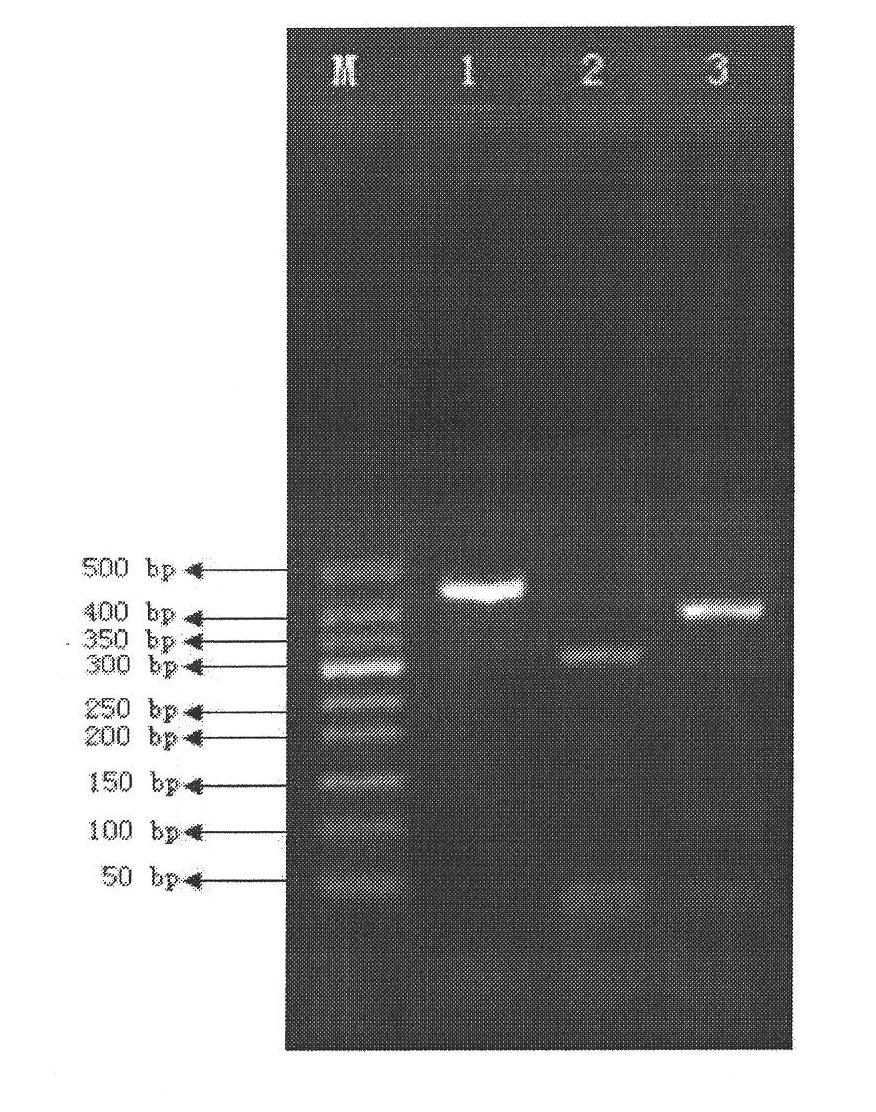
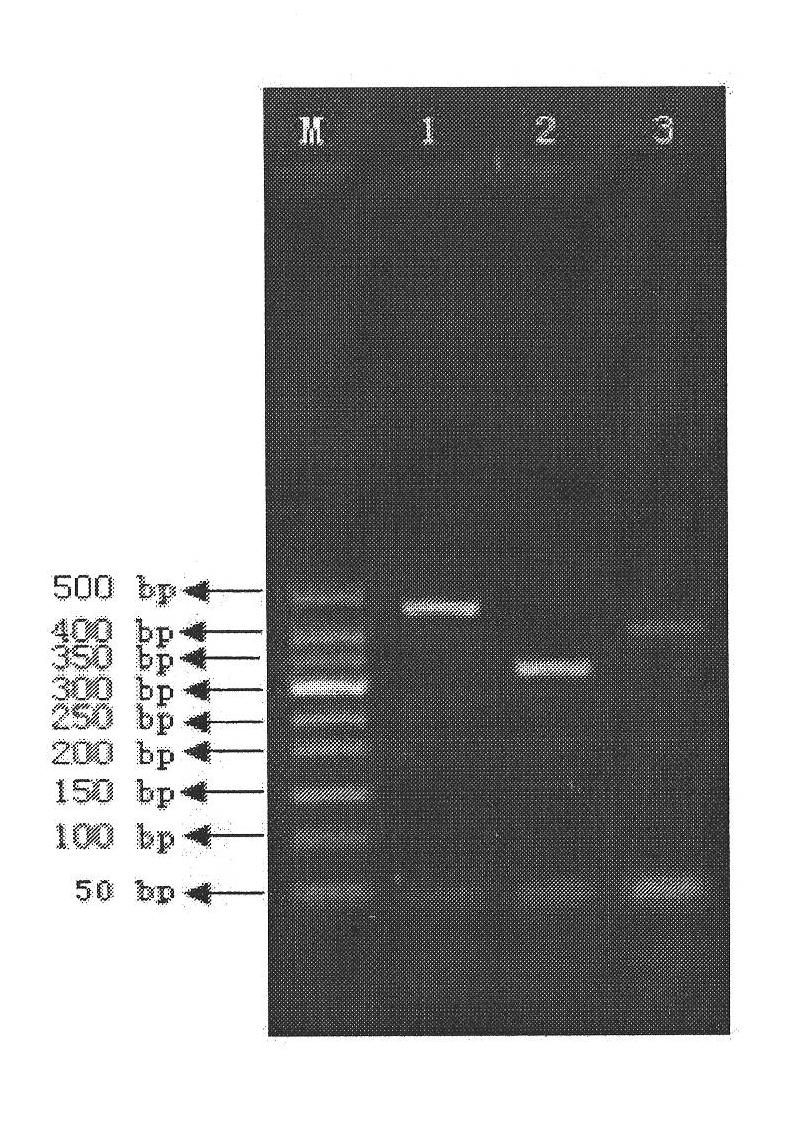
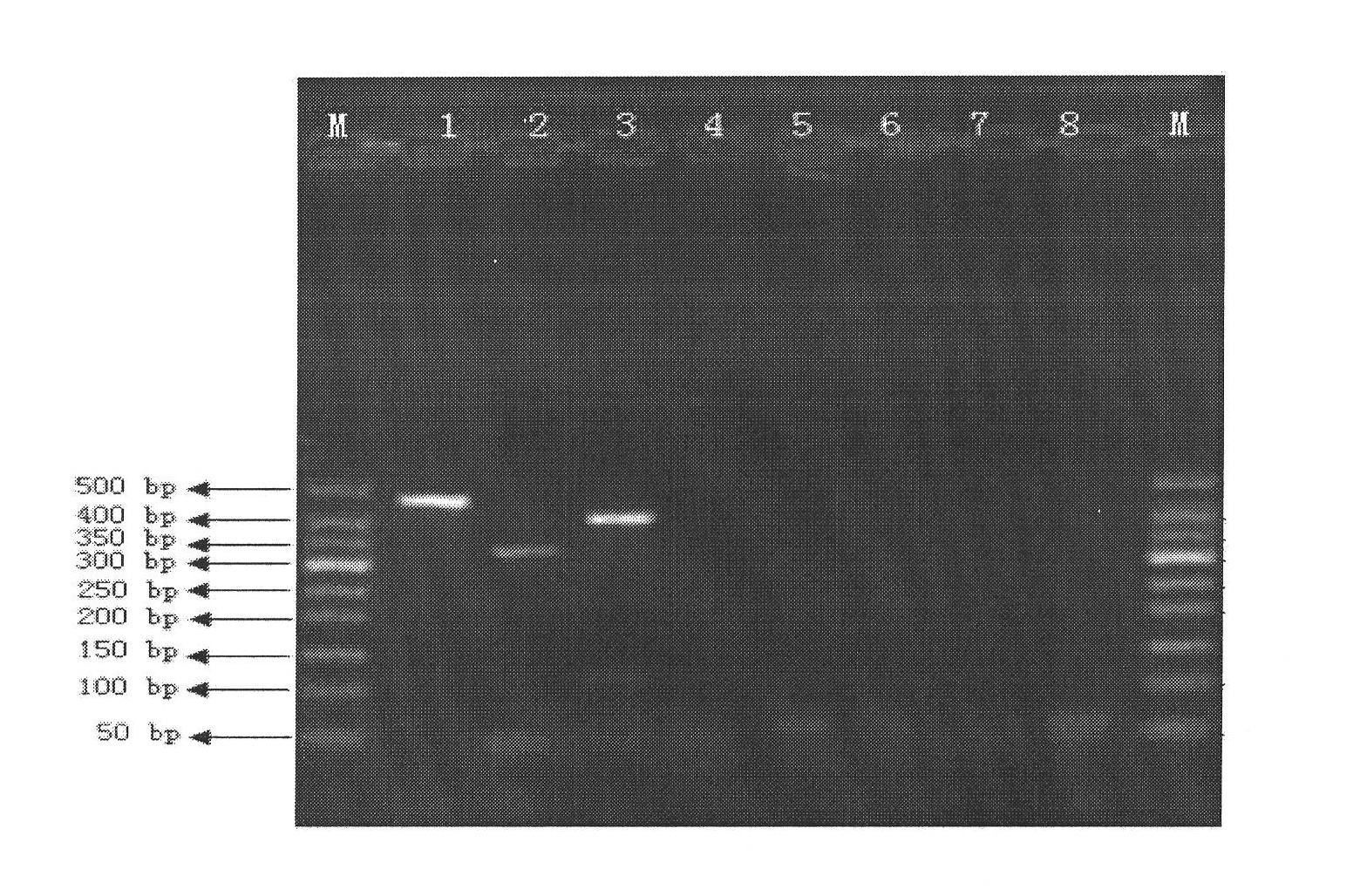
[0082] gyrB was amplified by PCR using the genomes of Legionella mikk-Dade, Legionella bozemannii and Legionella longbeach as templates. In the present invention, degenerate primers are designed according to the conserved region of gyrB, and the sequence of amplifying its gyrB degenerate primers is: upstream primer (5'-WCVGGTYTGCAYCAYATG'), downstream primer (5'-GTCTGBGAKGARAAYTTVGG'); The PCR reaction program is as follows: pre-denaturation at 94°C for 5 minutes; then denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, and extension at 72°C for 1 minute, thus performing 30 cycles; finally, extending at 72°C for 5 minutes to obtain PCR The size and specificity of PCR products were detected by 0.8% agarose gel electrophoresis. Combine 2 tubes of PCR products, and use the UNIQ-10 Column DNA Gel Recovery Kit from Shanghai Sangon B...
Embodiment 3
[0083] Example 3 : Sequencing and splicing of gyrB gene
[0084] The gel-cut and purified fragments were bidirectionally sequenced with an ABI377 automatic DNA sequencer, and the Staden Package was used for sequence splicing to obtain the sequence of the entire gyrB gene (SEQ ID NO: 1-3).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com