Rice lateral root control gene osiaa11 and its application
A gene and lateral root technology, applied in the fields of application, genetic engineering, plant genetic improvement, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1, Screening and phenotype of mutants
[0042] Using the Kasalath mutant library induced by EMS as the mutant screening object, M 2 Substitute seeds were rinsed with distilled water and diluted with 0.6% (v / v) HNO 3 Dormancy-breaking treatment for 16 hours, germination in the dark at 37°C until white. Sow the lubai seeds on the nylon mesh floating on the rice culture solution (the formula of the culture solution is the standard formula of the International Rice Institute), and cultivate it for 7 hours at a temperature of about 30 / 22°C (day / night) and 12 hours of light. Day, with the change of lateral roots (including length, quantity, etc.) as screening criteria for the screening of mutants, from which a mutant without lateral roots was screened ( figure 1 A-F), ie, rice lateral root development-deficient mutant Osiaa11.
Embodiment 2
[0043] Embodiment 2, gene localization
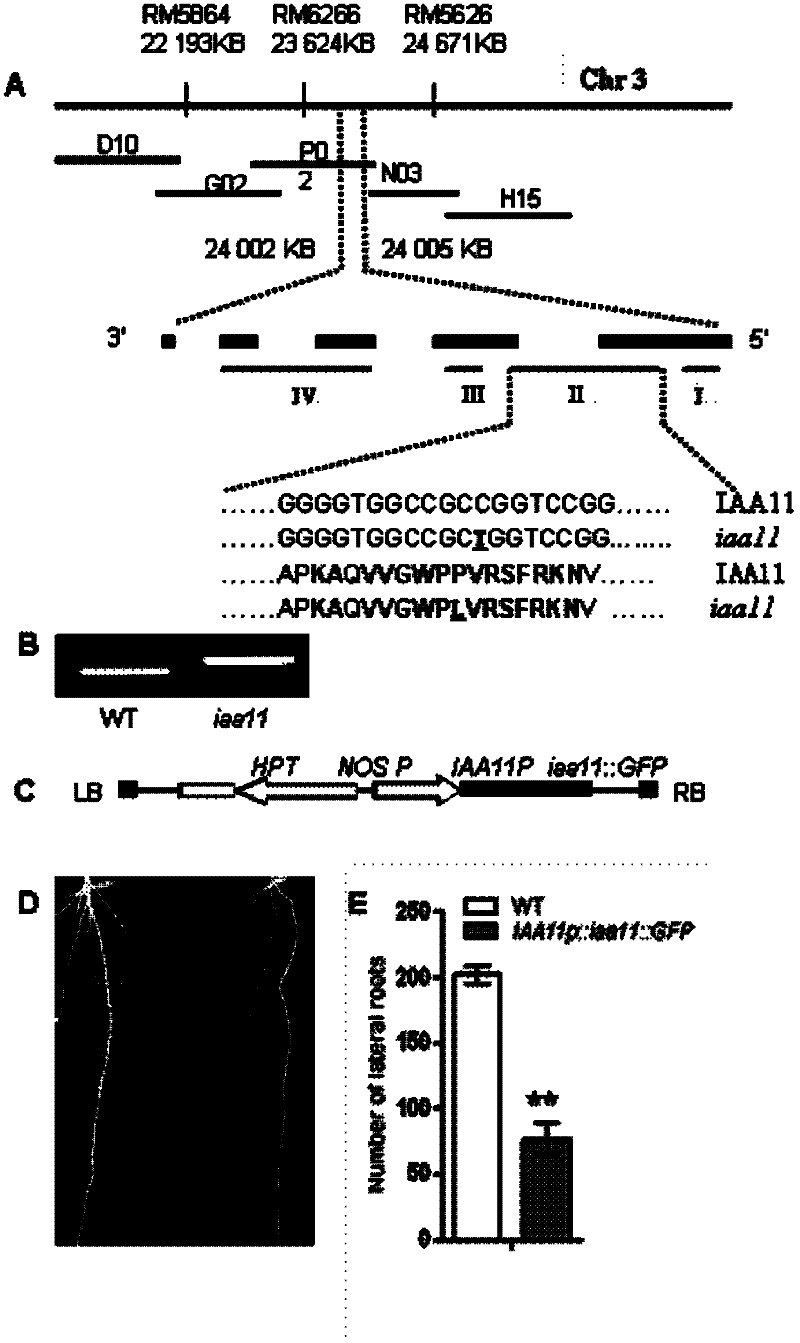
[0044] f 2 The mapping population was obtained by crossing the homozygous (Osiaa11) as the female parent and the wild-type japonica cultivar Nipponbare as the male parent, and a total of 186 F 2 Individuals, using the rapid extraction method of rice trace DNA to extract genomic DNA for gene mapping from rice leaves. Take about 2cm young rice leaves, freeze them with liquid nitrogen, grind the leaves into powder in a 1.5ml centrifuge tube, extract the total DNA, and dissolve the obtained DNA in 200 μl sterile water. 2 μl of DNA sample was used for each SSR and STS reaction.
[0045] 1), in the preliminary localization test of OsIAA11 gene, for 30 F 2 Individuals were subjected to SSR analysis. According to the published molecular genetic maps created by japonica and indica rice, select SSR primers that are approximately evenly distributed on each chromosome, perform PCR amplification according to known reaction conditions, and then s...
Embodiment 3
[0059] Example 3, gene prediction and sequence analysis
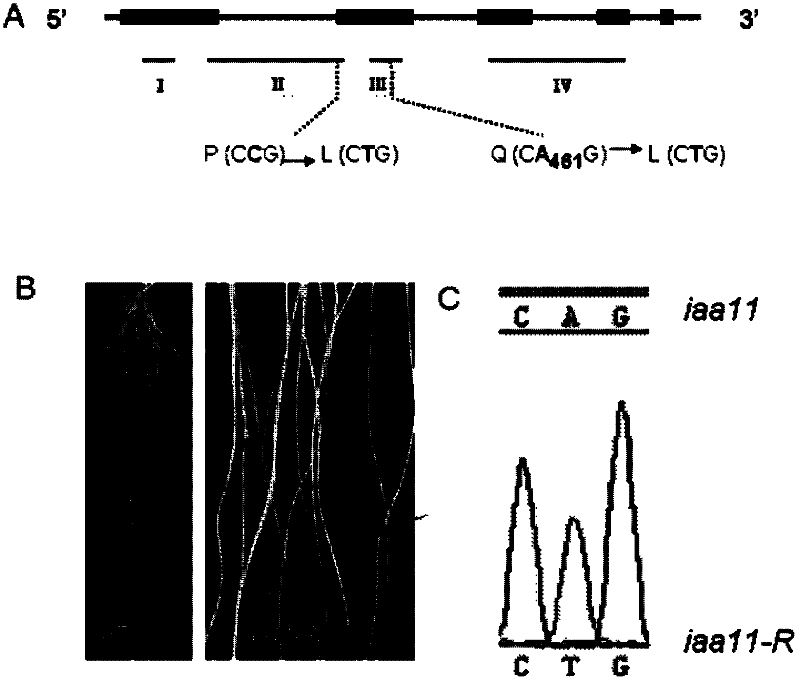
[0060] According to the results of fine mapping, the OsIAA11 gene is located within the range of RM6266 and RM5626 on the BAC clone OSJNBb0069P02 ( figure 2 A). According to the rice gene annotation information of TIGR (http: / / www.tigr.org / tdb / e2k1 / osa1 / ), the gene prediction analysis of the mapped chromosome segment shows that there are 2 AUX / IAA genes in this segment. Using Kasalath wild-type and Osiaa11 mutant cDNA as templates to amplify the two candidate genes, the amplified products were sequenced separately, and the sequencing results were compared and analyzed. It was found that the base C at the 317th bp after the start codon ATG of the IAA11 gene was Mutated into a T, resulting in an amino acid change from proline to leucine, this point mutation has been verified by dCAPs ( figure 2 B).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com