Escherichia coli expression vector capable of controlling self-cracking of host bacterium
An expression vector and Escherichia coli technology, applied in the fields of microorganisms and genetic engineering, can solve the problems of high energy consumption, inactivation of target products, expensive mechanical equipment, etc., and achieve the effect of facilitating product recovery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1: Codon and endonuclease recognition site optimization of T4 lysozyme gene
[0058] 1) Site-directed mutation of the codon AGA located at both ends of the gene
[0059] The full sequence of the original T4 lysozyme structural gene is SEQ ID NO:3. Analysis of the structural gene of T4 lysozyme showed that three of the codons encoding arginine were AGA, which were codons that greatly affected gene expression in Escherichia coli and would seriously reduce the expression level of the gene. One of these three codons is near the 5' end of the gene, and the other is near the 3' end of the gene. These two codons can be directly mutated when amplifying the gene using the PCR method. It contains a Mlu I recognition site downstream of the AGA codon near the 3' end. Mlu I is a commonly used enzyme cutting site in genetic engineering. If it is retained in the gene, it may cause inconvenience to the construction of expression vectors and the application of vectors. The...
Embodiment 2
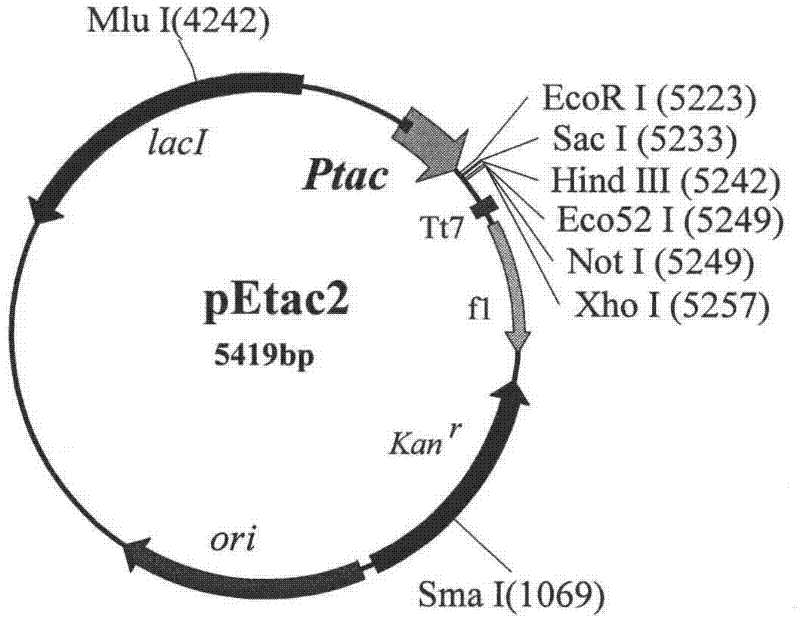
[0074] Embodiment 2: construction of plasmid pEtac2
[0075] Plasmid pEtac was double-digested with BglII and BamH I, and a fragment of about 5.4 kb was recovered by electrophoresis separation of the digested product. After purification, it was self-ligated, and the ligated product was transformed into E.coli JM109, and the transformants were screened on a kanamycin plate. Four transformants were randomly selected to extract plasmids and digested with Bgl II. The results showed that none of the four plasmids could be cut by Bgl II, which conformed to the characteristics of pEtac2. BglII and BamH I are homologous enzymes, which produce the same sticky ends after digestion, and can be annealed to each other, but the original restriction site disappears after connection, and neither enzyme can cut a new site. There is an Nco I restriction site between the Bgl II and BamH I sites in the original pEtac vector, and Nco I will not be able to cut the new plasmid after the fragment bet...
Embodiment 3
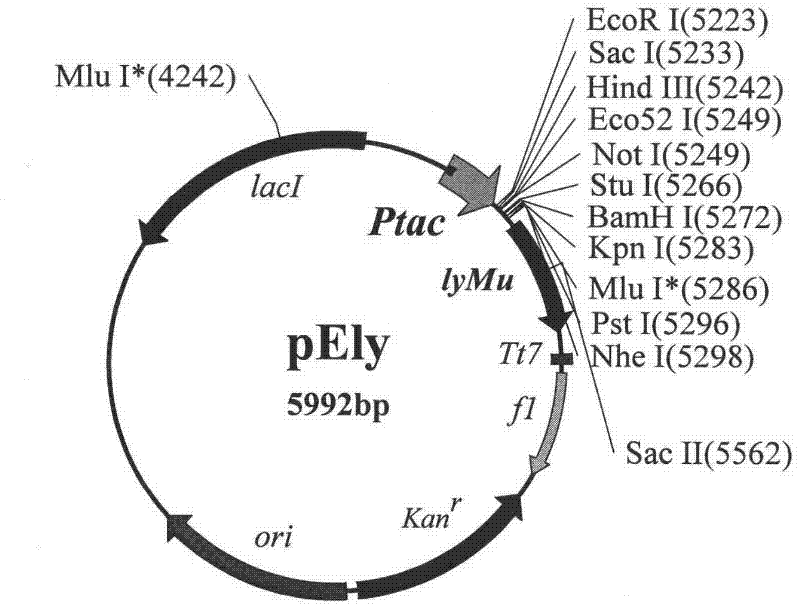
[0077] The construction of embodiment 3pEly expression vector
[0078] The lyMu gene fragment of about 580 bp was recovered by electrophoresis after the plasmid pMD-lyMu was digested with Sal I. Plasmid pEtac2 was digested with Xho I and ligated with the above-mentioned recovered fragment, the ligated product was transformed into Escherichia coli JM109, and transformants were screened on a kanamycin plate. A total of 20 transformants were selected to extract plasmids, and these plasmids were used as templates to carry out PCR with the following primers.
[0079] Ply 03: 5'-TTACGTATAGATGAACGTC-3' (SEQ ID NO: 8)
[0080] Pkan: 5'-GTTTCATTTGATGCTCGATG-3' (SEQ ID NO: 9)
[0081] The PCR amplification conditions were denaturation at 95°C for 5 minutes, 25 cycles of amplification reaction (94°C for 50 s, 56°C for 60 s, 72°C for 1 min), and extension at 72°C for 5 min. The plasmid extracted from one of the transformants was a template PCR to obtain an amplified fragment of about 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com