Method for cloning deoxyribonucleic acid (DNA)
A DNA cloning and carrier technology, applied in the field of DNA cloning, can solve problems such as false positives, difficulty in connecting large fragments of DNA, and increased screening workload, etc., and achieve the effects of short reaction time, improved cloning success rate, and high positive cloning rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] In this example, the human gene PRAK was inserted into the BamHI and XhoI sites of the viral vector pBOB. The specific operations are as follows:
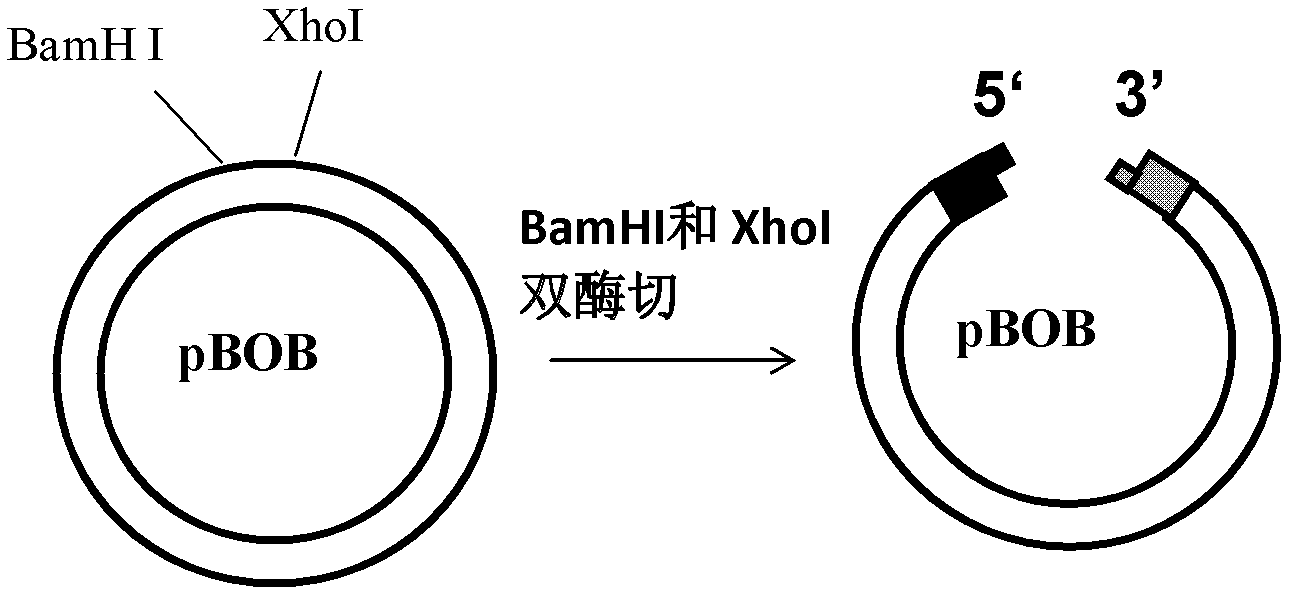
[0049] 1) Digest the circular vector pBOB with BamHI and XhoI to obtain a linear vector with 5'protruding ends at both ends ( figure 1 );
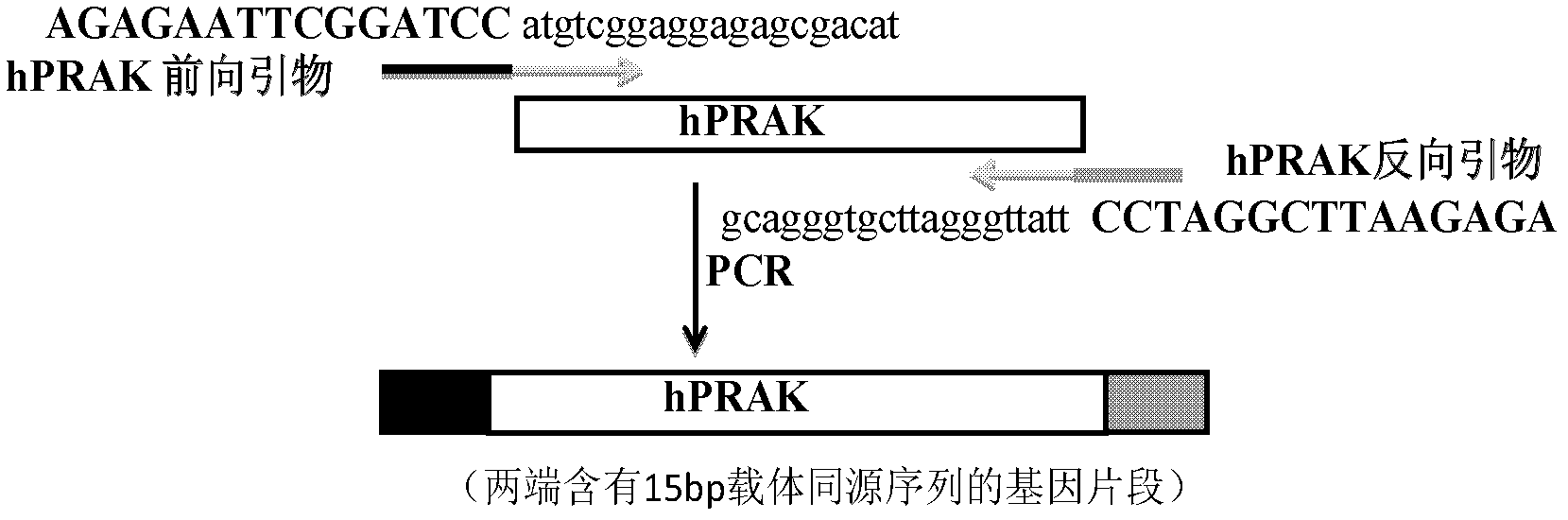
[0050] 2) Design two primers based on the sequence of the target gene hPRAK and pBOB at the upstream 5'end of BamHI and the downstream 3'end of XhoI to amplify the target gene. The two primers are 35 bp in length. The 5'end contains the 15bp vector pBOB homologous sequence (indicated by capital letters), and the 3'end contains the 20bp target gene hPRAK homologous sequence (indicated by lowercase letters).
[0051] hPRAK forward primer sequence: 5'-AGA GAA TTC GGA TCC atgtcggaggagagcgacat-3';
[0052] (The uppercase letters are the homologous sequence at the upstream 5'end of the pBOB vector BamHI, and the lowercase letters are the forward amplification primers at the 5'end of the target gene)
...
Embodiment 2
[0067] The basic steps are the same as in Example 1, the difference is:
[0068] 1) The insertion site of the target gene in the vector pBOB is SmaI. The linear vector generated by the restriction enzyme SmaI in step 1 has blunt ends at both ends;
[0069] 2) The two primers designed by step 3 for PCR amplification of hPRAK are 35 bp in length. The 5'end contains 15 bp vector pBOB homologous sequence (indicated by capital letters), and the 3'end contains 20 bp target gene hPRAK homologous sequence (using Lowercase letters).
[0070] hPRAK forward primer sequence: 5’-TCC AAT ATT CCC GGG atgtcggaggagagcgacat-3’;
[0071] (The uppercase letters are the homologous sequence at the 5'end of the upstream of pBOB vector SmaI, and the lowercase letters are the forward amplification primers at the 5'end of the target gene)
[0072] hPRAK reverse primer sequence: 5'-TGG CTC GAG CCC GGG ttattgggattcgtgggacg-3'.
[0073] (Uppercase letters are the homologous sequence at the downstream 3'end of pBOB ...
Embodiment 3
[0075] The basic steps are the same as in Example 1, except that: the length of the primer designed in step 3 can be changed, and the length of the homologous sequence of the 5'end of the primer can be between 8-20 bp.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com