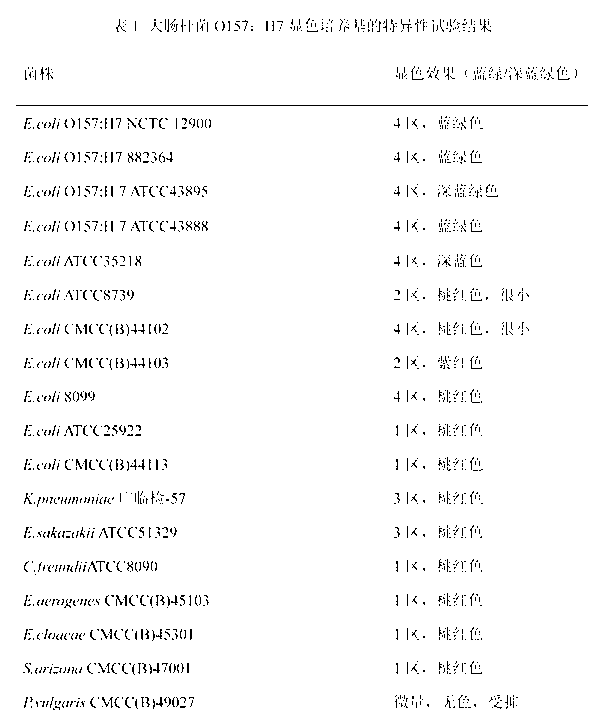
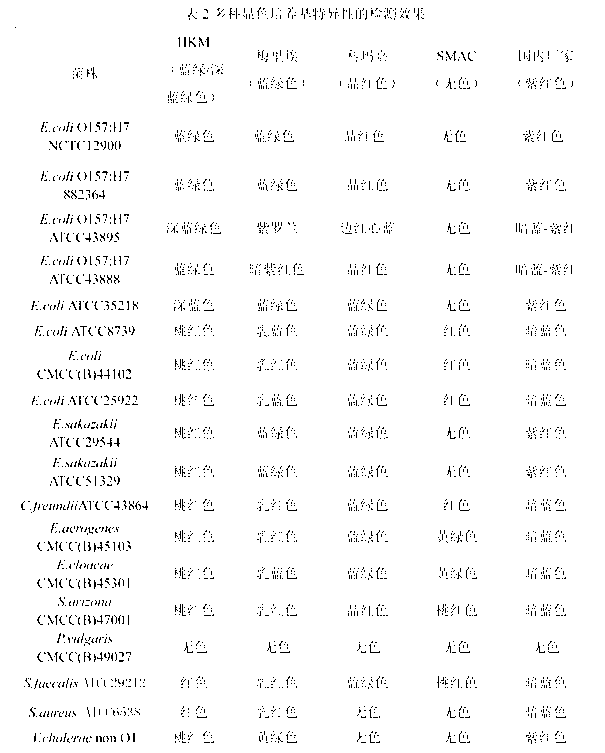
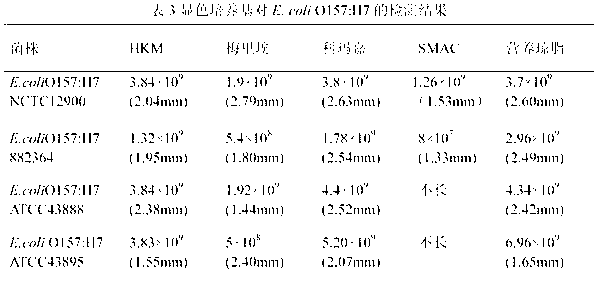
Chromogenic medium used for detecting esherichia coli O157:H7
A chromogenic medium, O157 technology, applied in the direction of microbe-based methods, microbiological determination/inspection, microbiology, etc., can solve problems such as false positives, and achieve the effects of short detection cycle, strong operability, and high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032]A chromogenic medium for detecting Escherichia coli O157:H7, each 1000mL medium contains 14g of agar, 10g of peptone, 4g of beef extract powder, 5g of sodium chloride, 15g of sorbitol, 5g of inositol, and 0.01g of neutral red , 5-bromo-4-chloro-3-indole-β-galactoside 0.1g, 5-bromo-6-chloro-3-indole-β-glucuronide 0.08g, isopropyl-β- D-thiogalactopyranoside (IPTG) 0.1g, ox bile salt 1.5g, potassium tellurite 2mg, cefixime 0.04mg.
Embodiment 2
[0034] A chromogenic medium for detecting Escherichia coli O157:H7, each 1000mL medium contains 12g of agar, 12g of peptone, 3g of beef extract powder, 7g of sodium chloride, 15g of sorbitol, 4g of inositol, and 0.005g of neutral red , 5-bromo-4-chloro-3-indole-β-galactoside 0.05g, 5-bromo-6-chloro-3-indole-β-glucuronide 0.3g, IPTG 0.05g, ox bile Salt 1g, potassium tellurite 3mg, cefixime 0.01mg.
Embodiment 3
[0036] A chromogenic medium for detecting Escherichia coli O157:H7, each 1000mL medium contains 15g of agar, 20g of peptone, 7g of beef extract powder, 6g of sodium chloride, 20g of sorbitol, 7g of inositol, and 0.03g of neutral red , 5-bromo-4-chloro-3-indole-β-galactoside 0.2g, 5-bromo-6-chloro-3-indole-β-glucuronide 0.05g, IPTG 0.2g, ox bile Salt 5g, potassium tellurite 5mg, cefixime 0.06mg.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com