Codon optimized fatty acid desaturase gene sequence and application thereof
A fatty acid dehydrogenase and codon optimization technology, applied in application, genetic engineering, plant genetic improvement and other directions, can solve problems such as unfavorable direct expression, different preferences, etc., achieving good applicability, simple process, and solving ratio imbalance. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1: In this embodiment, the cell strains that stably express the FatI gene sequence that promotes the conversion of n-6 fatty acids in cells to n-3 fatty acids are cultivated according to the following steps:
[0038] 1.1 CHO cell culture: after the routine resuscitation of the CHO cell line, the DMEM medium containing 10% fetal bovine serum, 100U / ml penicillin and 100mg / L streptomycin sulfate (the present invention has no special instructions, all use this medium) Cultivate in an incubator at 37°C with 5% CO2 environment, and use for cell transfection when the cell confluency is about 85%.
[0039] 1.2 Vector construction: According to the nematode sequence (ACCESSION NM_001028389) in GenBank, the codon-optimized FatI gene sequence for mammals was synthesized by chemical synthesis, and a sequence GTTGCGGCCGCCACC was added before the start codon ATG, which contained The Kozak sequence that is beneficial to gene expression, and the restriction site of HindIII a...
Embodiment 2
[0042] Embodiment 2: In this embodiment, the cell line detection and its fatty acid analysis are carried out according to the following steps:
[0043] 2.1 Preliminary detection of positive cells by genomic PCR: the cells left on the original culture plate during the cloning and amplification culture process continued to be cultured, and when the cells covered the culture plate, they were digested with trypsin to collect the cells, and the genome of the cells was extracted by the genome extraction kit, and RT-PCR Preliminary detection of whether the exogenous gene has entered the cell, the primers used are FatI-F: 5′-CAT GGT CGC TCA CTC CAG C-3′, FatI-R: 5′-GGT ACC TTA CTT AGC TTT GGC CTT TTC-3′; Reaction conditions: 94°C for 5 min, 94°C for 1 min, 60°C for 30 sec, 72°C for 1 min; 30 cycles, 72°C for 5 min; the positive cell lines detected by agarose gel electrophoresis were used for the next experiment.
[0044] 2.2 Southern Blot detection: The vector pCDNA3.1(+)-FatI constru...
Embodiment 3
[0047] Embodiment 3: This embodiment analyzes the optimization results as follows:
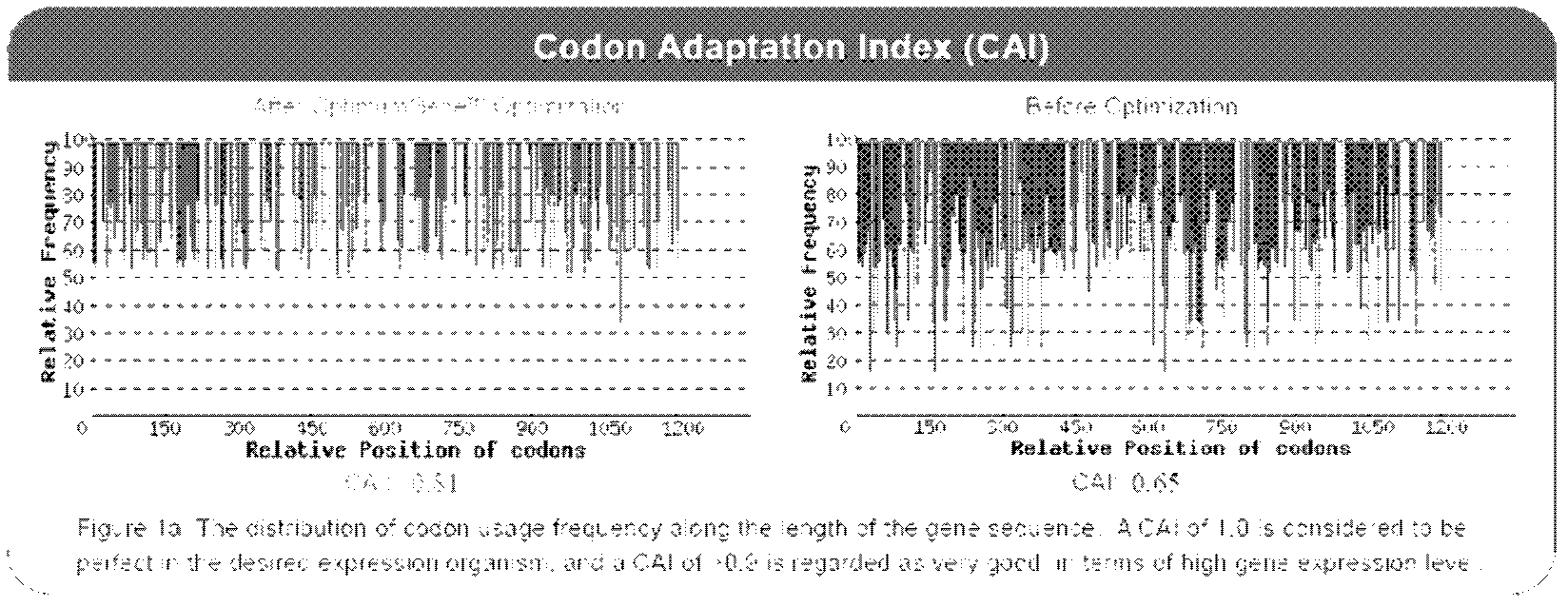
[0048] 3.1 Codon optimization, gene synthesis and construction of expression vector: According to the codon preference of mammals, codon optimization was carried out on the FatI gene sequence of nematodes; the CAI of the optimized sequence was increased from 0.65 to 0.81 (attached figure 2 ), it is generally believed that when the CAI is greater than 0.9, it is very conducive to gene expression, and a sequence of GTTGCGGCCGCCACC is added before the start codon ATG, which contains the Kozak sequence that is beneficial to gene expression, and HindIII is introduced in its upstream and downstream, respectively, The cut site of KpnI restriction endonuclease and the corresponding protective bases; the comparison between the optimized sequence and the original sequence is as attached figure 1 , the optimized sequence part is shown in red; the optimized sequence was synthesized by chemical synthesis...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com