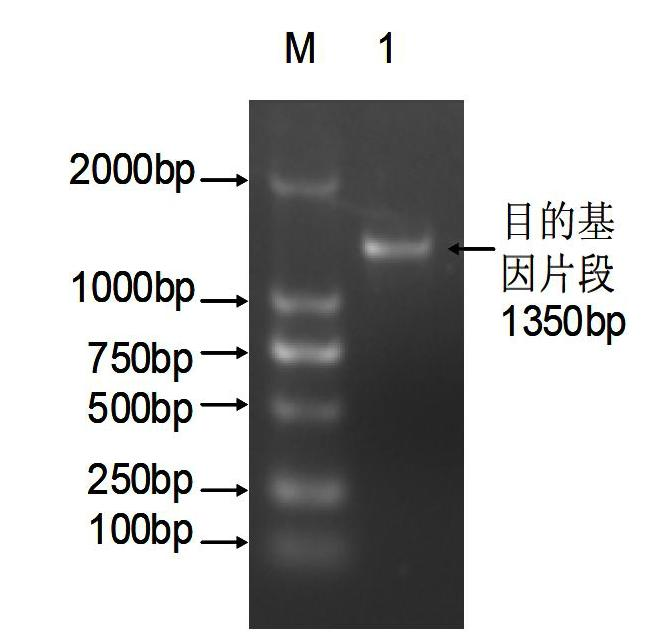
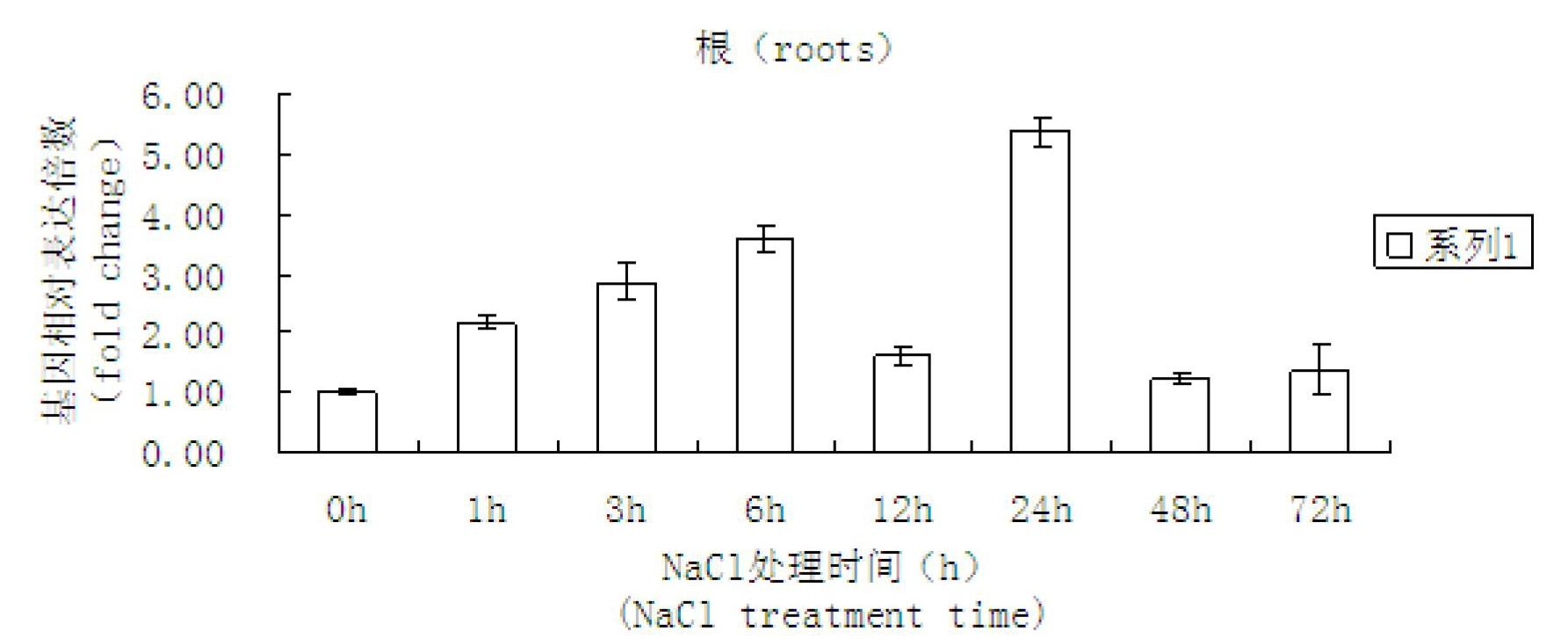
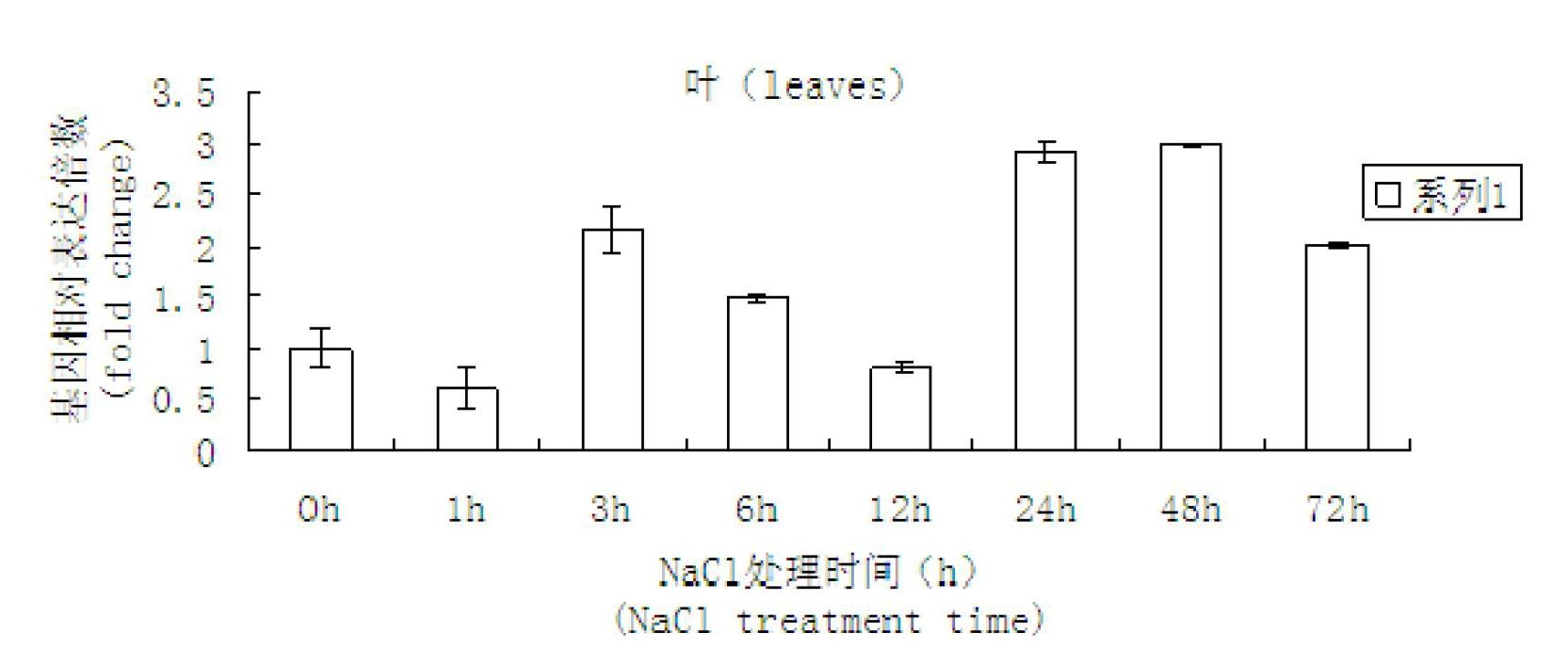
Cotton salt-tolerant gene GarCIPK for improving plant salt tolerance
A technology of salt tolerance and cotton, applied in the field of biological genetic engineering, can solve the problem of less research on cotton, and achieve the effect of improving salt tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Implementation example 1, GarCIPK Gene acquisition
[0020] 1.1 Extraction of RNA
[0021] extract RNA
[0022] (1) Take 0.5g of fresh cotton tissue, add 0.1g of cross-linked polyvinylpyrrolidone (PVPP), fully grind to powder in liquid nitrogen, quickly transfer the frozen powder into a 10ml centrifuge tube, add 5ml of CTAB extract With 500 μL of 0.1M Tris-HCl of pH 8.0, bathe in water at 65°C for 20 minutes, and mix well by inverting halfway;
[0023] (2) Add an equal volume of chloroform to mix thoroughly, and let stand in an ice bath for 10 minutes;
[0024] (3) Centrifuge at 10,000 rpm for 20 minutes at 4°C. Divide into four 1.5ml centrifuge tubes;
[0025] (4) Aspirate the supernatant, add 1 / 3 volume of 8M LiCl, mix well, and incubate at -70°C for 30min or -20°C overnight;
[0026] (5) Centrifuge at 10,000 rpm for 20 minutes at 4°C. Discard the supernatant, wash twice with 70% ethanol, dry the precipitate and dissolve it in 30 μL DEPC water;
[0027] (6)...
Embodiment 2
[0076] Embodiment 2, the construction of plant expression vector
[0077] 2.1 Construction of pCAMBIA2301-CaMV35S-GarCIPK plant expression vector
[0078] Plant expression vector pCAMBIA2301-CaMV35S (purchased from Biovector Science Lab) plasmid, select BamHI and KnpⅠ to digest pCAMBIA2301 and the target gene fragment respectively, recover the large vector fragment and the target gene fragment, connect with T4 ligase and transform Escherichia coli DH5α to sense state cells, and the plant expression vector with the target gene can be obtained after the recombinant is identified.
[0079] Enzyme digestion of pCAMBIA2301 plasmid and target gene fragment
[0080] The plasmid double enzyme digestion system is as follows:
[0081] 10×K buffer 5μL
[0082] 2301 plasmid 25μL
[0083] BamHI 0.5 μL
[0084] KnpI 0.5 μL
[0085] sterile ddH 2 O 19 μL
[0086] Enzyme digestion at 30°C for 5h. After the double enzyme digestion, the agaro...
Embodiment 3
[0111] Embodiment 3, preparation and transformation of Agrobacterium competent
[0112] 3.1 Preparation of Competent Agrobacterium EHA105
[0113] (1) Pick a single colony of EHA105, inoculate it in 5ml LB liquid medium, and culture overnight at 28°C with shaking at 200rpm until the OD600 value is 0.4;
[0114] (2) Inoculate in 400-500ml LB medium (in a 1L Erlenmeyer flask) at a ratio of 1:100, shake the bacteria until the OD600 is 0.6-0.8, and bathe in ice for 10 minutes;
[0115] (3) Collect the bacterial liquid in a pre-cooled 50ml centrifuge tube, centrifuge at 5000rpm at 4°C for 5min;
[0116] (4) Discard the supernatant, and use 10ml pre-cooled 0.1MCaCl for precipitation 2 Fully suspend, 4°C, 5000rpm, centrifuge for 5min;
[0117] (5) Add 4ml (depending on the number of bacteria) to pre-cool the solution containing 15% 0.1mol / L CaCl2, gently suspend the cells, and place them on ice for a few minutes to form a competent cell suspension;
[0118] (6) Divide into 100μL-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com