Method for quantitative determination of escherichia coli RNA, and specialized standard substance and application thereof
A quantitative detection technology for Escherichia coli, which is applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc. It can solve the problems of not considering the reverse transcription efficiency, underestimating the RNA content, etc., and achieve a linear detection range Wide, simple preparation method, good purity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, the synthesis of primer and the preparation of standard
[0039] 1. Primer design and synthesis
[0040] The genomic DNA of Escherichia coli was analyzed, the partial sequence of uidA gene was selected, and two pairs of primers (primer pair A and primer pair B) were designed according to the sequence.
[0041] Primer pair A is composed of uidA-T7-F and uidA-T7-R, and the target sequence is the double-stranded DNA molecule shown in sequence 1 of the sequence listing, which is 921bp.
[0042] uidA-T7-F (sequence 2): 5'-taatacgactcactataggggcgttacaagaaagcc-3';
[0043] uidA-T7-R (SEQ ID NO: 3): 5-gcatctcttcagcgtaagggtaatgcga-3'.
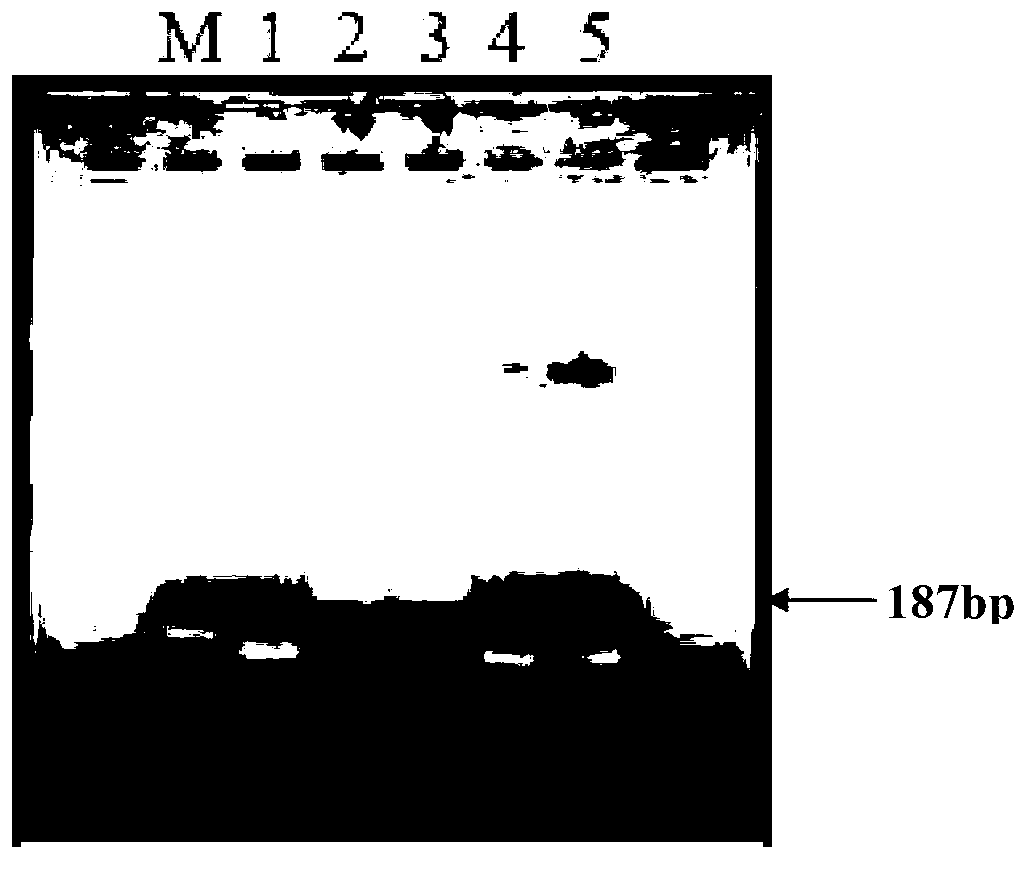
[0044] Primer pair B is composed of uidA-F and uidA-R, and the target sequence is the double-stranded DNA molecule shown in the 257th to 443rd nucleotides from the 5' end of sequence 1 in the sequence listing, which is 187bp.
[0045] uidA-F (sequence 4): 5'-cgatgtcacgccgtatgttatt-3';
[0046] uidA-R (SEQ ID NO: 5): 5'-ggtgtagag...
Embodiment 2
[0057] Embodiment 2, gradient dilution of standard substance
[0058] Method for detecting RNA copy number: Use an ultra-micro nucleic acid protein analyzer (NanoDrop ND-2000C, the United States) to measure the RNA concentration, and then calculate the RNA copy number according to the following formula:
[0059]
[0060] 6.02×10 23 is Avogadro's constant; 340 (Da) is the relative molecular mass of one base of RNA.
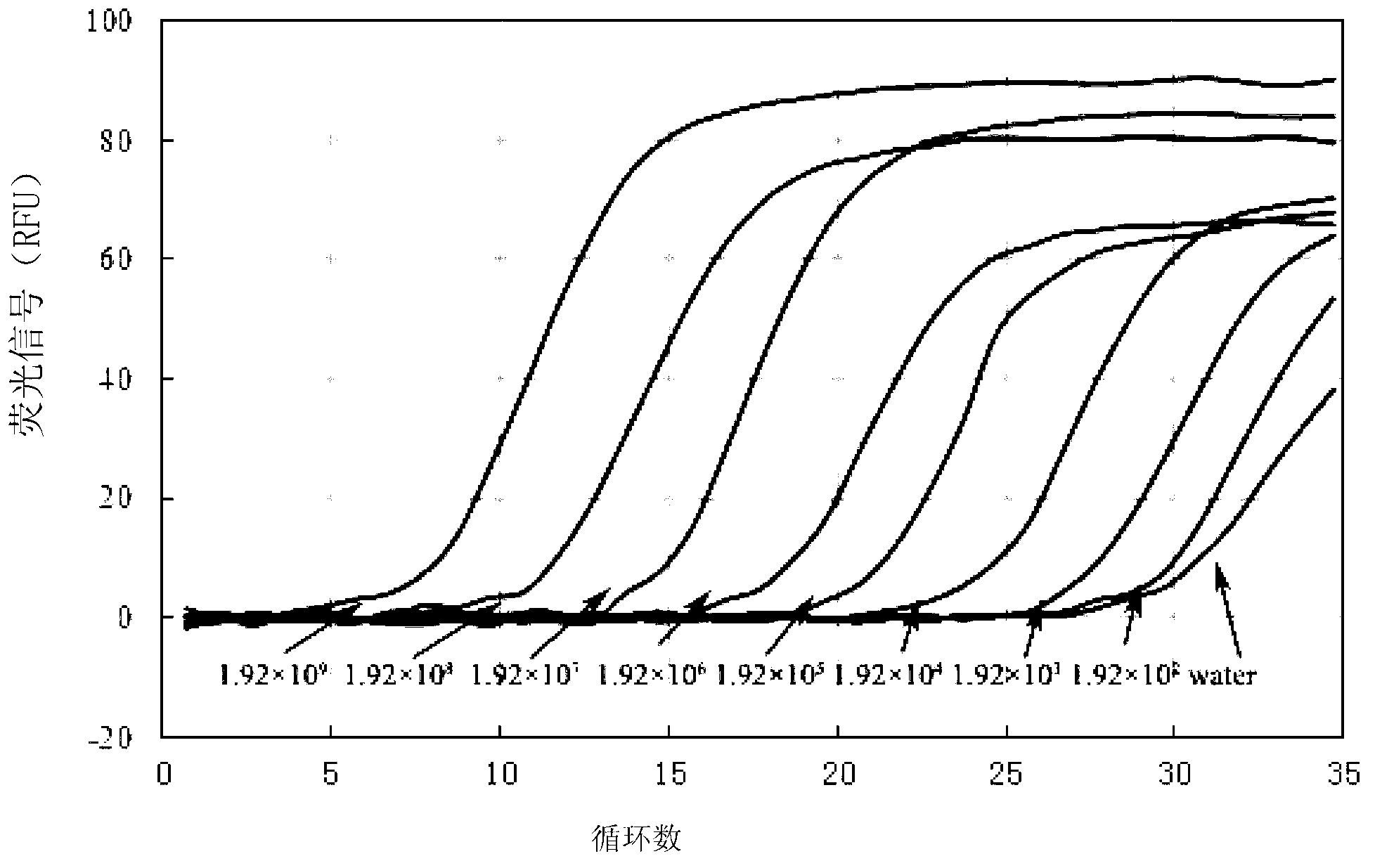
[0061] The standard substance prepared in Example 1 was sequentially diluted with sterile water to obtain dilutions 1 to 8, in which the concentration of RNA was: 9.6186×10 8 Copy number / μL, 9.6186×10 7 Copy number / μL, 9.6186×10 6 Copy number / μL, 9.6186×10 5 Copy number / μL, 9.6186×10 4 Copy number / μL, 9.6186×10 3 Copy number / μL, 9.6186×10 2 Copy number / μL and 9.6186×10 1 Copy number / μL.
Embodiment 3
[0062] Embodiment 3, the establishment of the method for the quantitative detection Escherichia coli living bacteria RNA
[0063] 1. Optimization of primer annealing temperature
[0064] 1. Using the cDNA after reverse transcription of the standard product obtained in Example 1 as a template, use the primers synthesized in Example 1 to perform PCR amplification on B.
[0065] The PCR reaction system is (20μL): 10×PCR Buffer 2μL, 25mM MgCl 2 1.6 μL, 1.6 μL of 10 mM dNTP, 1 μL of upstream primer (20 μM), 1 μL of downstream primer (20 μM), 2 μL of template (about 100 ng), 0.4 μL of Taq DNA polymerase (5U / μL), 10.4 μL of deionized water.
[0066] PCR reaction program: 95°C for 10min; 95°C for 30s, annealing for 20s, 72°C for 20s, 40 cycles; 72°C for 5min; store at 4°C after the reaction. 51°C, 52°C, 53.7°C, 54.4°C, and 55.5°C were used as annealing temperatures, respectively.
[0067] 2. Perform 2% agarose gel electrophoresis on the PCR amplified product of step 1, the results ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com