Specific primer and liquid chip for detecting polymorphism of NAT1 (N-acetyltransferase 1) gene
A gene polymorphism and detection solution technology, applied in the field of molecular biology, can solve problems such as unusable and unsuitable for practical applications, achieve consistent detection results, avoid uncertain factors, and improve detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The NAT1 gene polymorphism detection liquid chip described in this embodiment mainly includes:
[0028] 1. ASPE Primers
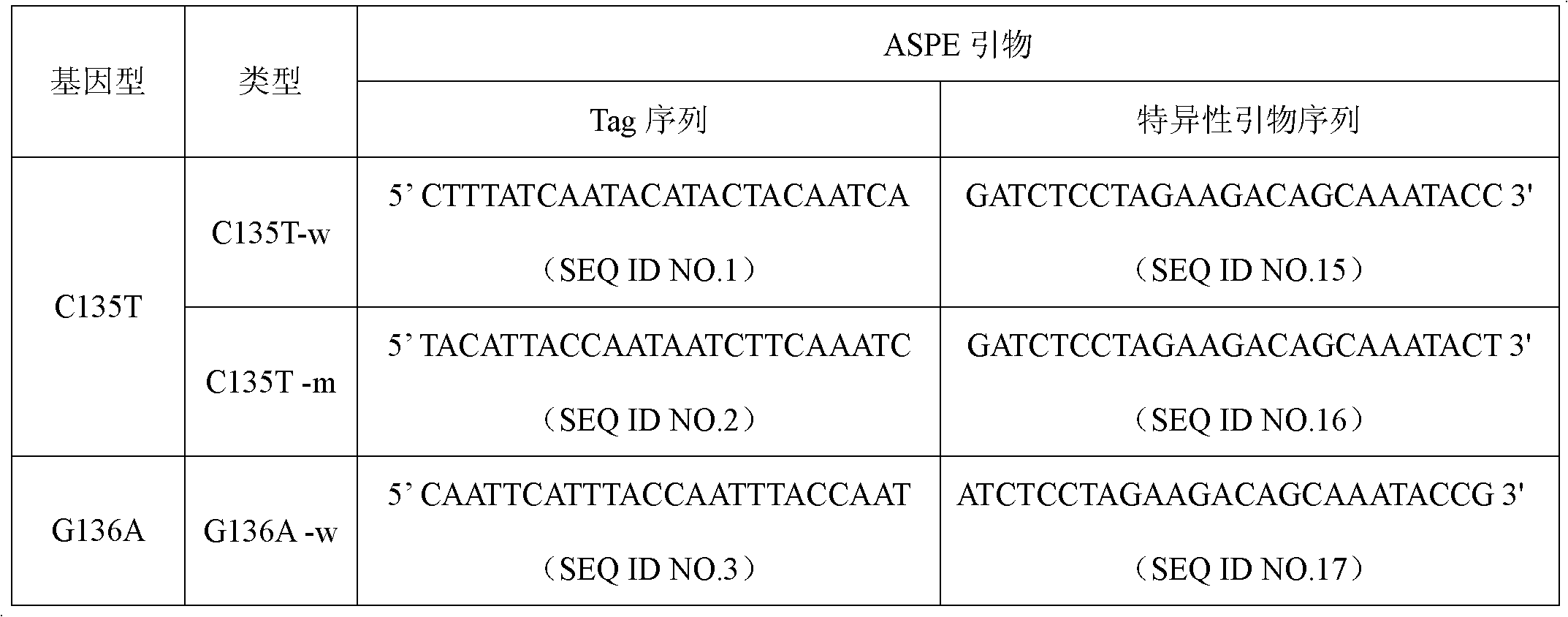
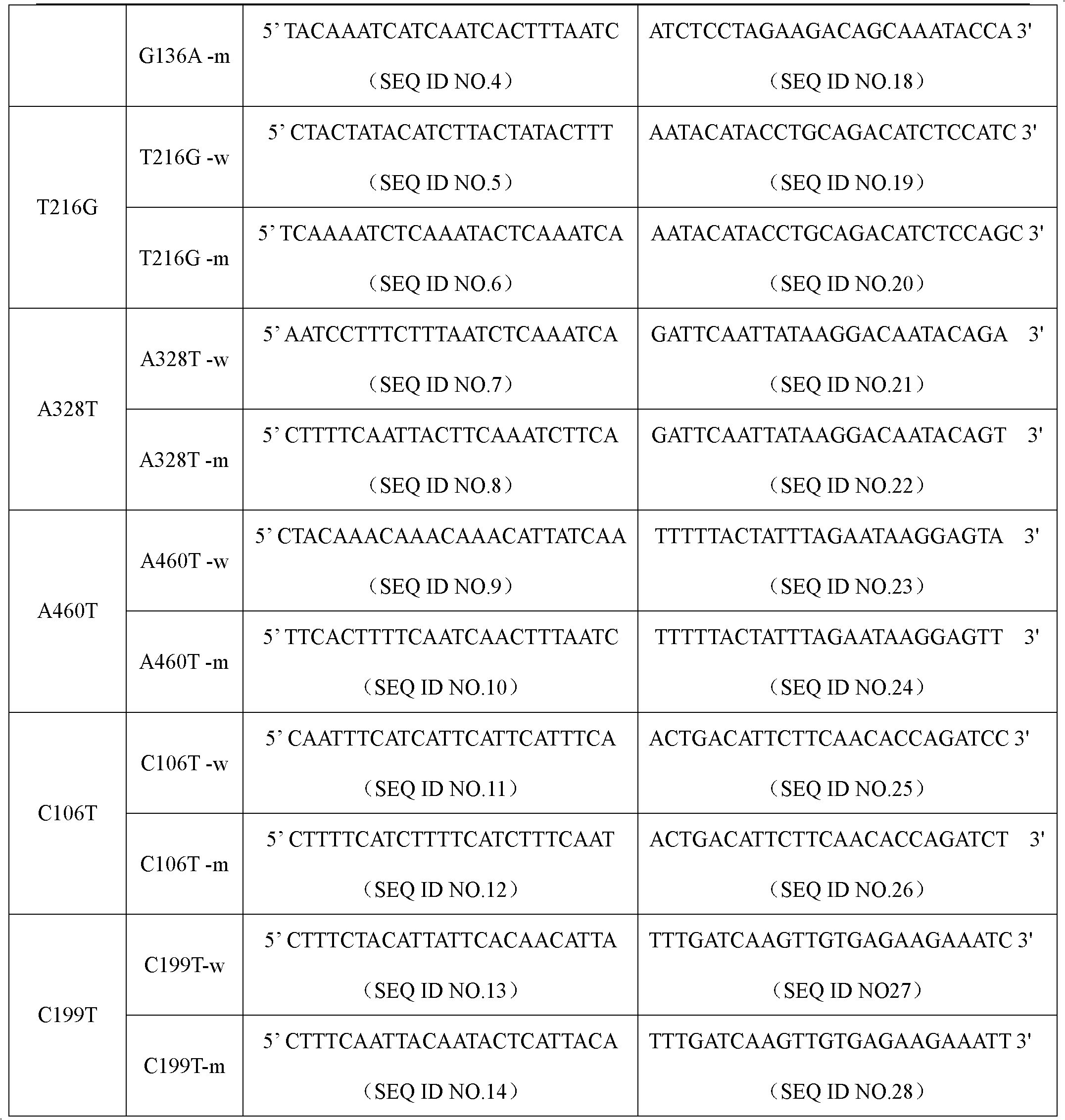
[0029] Specific primer sequences were designed for wild-type and mutant types of seven common genotypes of NAT1 gene, C135T, G136A, T216G, A328T, A460T, C106T and C199T. ASPE primers consist of "Tag sequence + specific primer sequence". ASPE primer sequences are shown in the table below:
[0030] Table 1 ASPE primer sequence (Tag sequence + specific primer sequence) of NAT1 gene
[0031]
[0032]
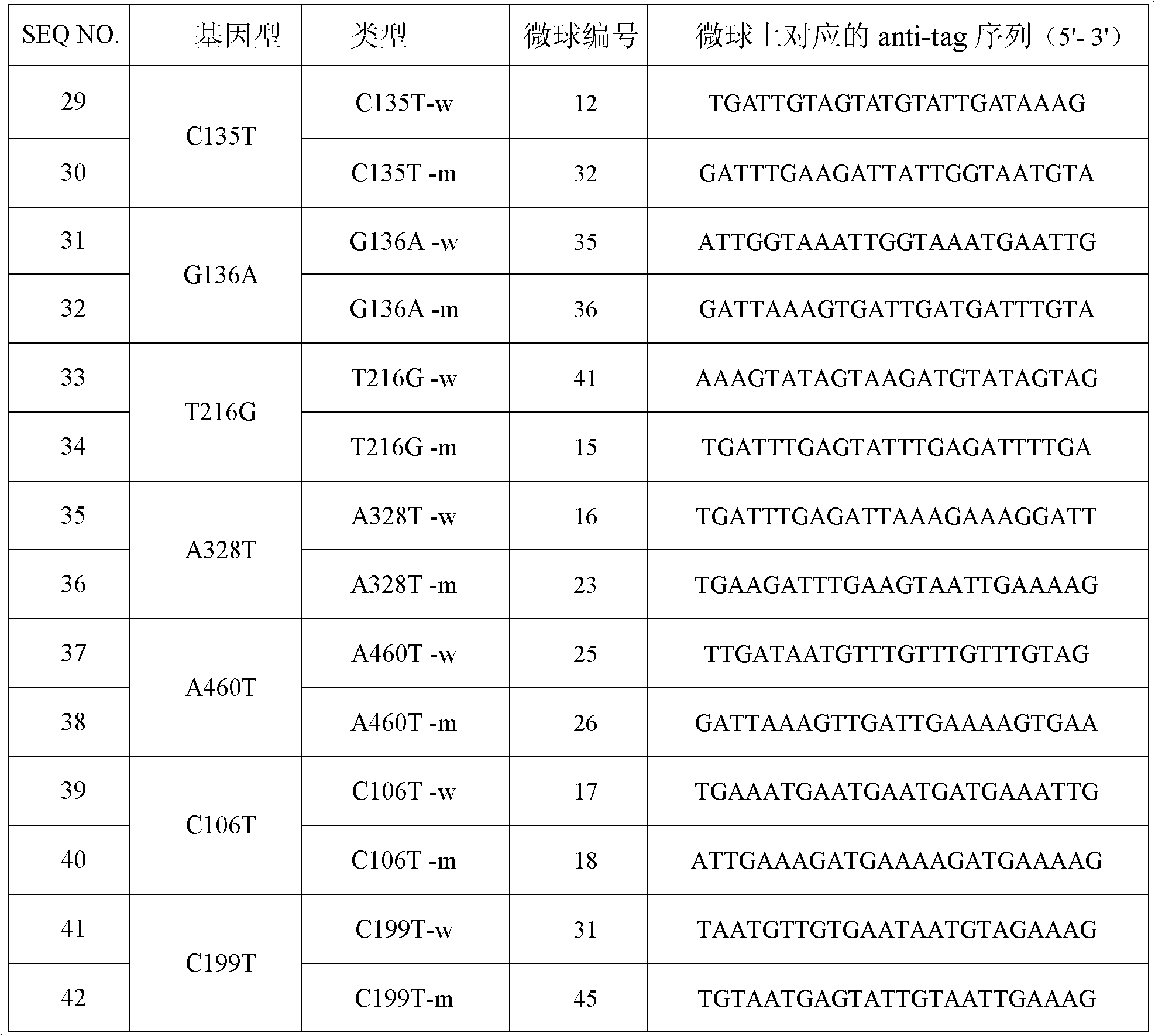
[0033] Each ASPE primer includes two parts, the 5' end is a specific tag sequence for the anti-tag sequence on the corresponding microsphere, and the 3' end is a mutant or wild-type specific primer fragment (as shown in the above table 1). All ASPE primers were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd. Each synthesized primer was prepared into a stock solution of 100 pmol / mL with 10 mmol / L Tris Buffer.
[0034] 2...
Embodiment 3
[0102] The liquid phase chip of embodiment 3 different ASPE primers detects the polymorphic site of NAT1 gene
[0103] 1. Design of liquid phase chip preparation (selection of Tag sequence and Anti-Tag sequence)
[0104] Taking the NAT1 gene G136A and A328T site mutation detection liquid chip as an example, the specific primer sequence of the 3' end of the ASPE primer was designed for the wild type and mutant type of G136A and A328T, and the Tag sequence at the 5' end of the ASPE primer was selected from SEQ ID NO.1-SEQ ID NO.14, correspondingly, the anti-tag sequence coated on the microsphere and complementary to the corresponding tag sequence is selected from SEQ ID NO.29-SEQ ID NO.42. The specific design is shown in the following table (Table 8). The synthesis of ASPE primers, microspheres coated with anti-tag sequences, amplification primers, detection methods, etc. are as described in Example 1 and Example 2.
[0105] Table 8 Design of liquid phase chip preparation
[...
Embodiment 4
[0116] Example 4 Selection of Specific Primer Sequences for NAT1 Gene Polymorphism Detection
[0117] 1. Design of liquid-phase chip preparation (selection of wild-type and mutant-specific primer sequences)
[0118] Taking the polymorphic site detection liquid chip of NAT1 gene T216G and A460T as an example, using the forward or reverse complementary sequence of the target sequence where the mutation site is located as a template, the wild type and mutant type of T216G and A460T were designed respectively The specific primer sequences at the 3' end of the ASPE primers include the preferred specific primer sequences and 2 alternative specific primer sequences in Example 1 of the present invention, as shown in Table 11. in, Inner bases are polymorphic sites.
[0119] Table 11 specific primer sequence
[0120]
[0121]
[0122] Taking the polymorphic site detection liquid chip of NAT1 gene T216G and A460T as an example, different specific primer sequences were selected ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com