Bacterium drug-resistant gene detection method, gene chip and kit
A technology for detecting kits and drug-resistant genes, which is applied in the field of biomedicine to achieve significant clinical practical value, fast detection speed, high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Using the kit of the present invention to detect drug resistance genes of common clinical drug-resistant bacteria
[0029] 1. Detection target and design of primers and probes
[0030] The detection targets of this embodiment include: Staphylococcus aureus and Enterococcus drug resistance genes in Gram-positive cocci, and Klebsiella pneumoniae, Escherichia coli, Enterobacter cloacae, Pseudomonas aeruginosa in Gram-negative bacilli Resistant genes of spp. and Acinetobacter baumannii.
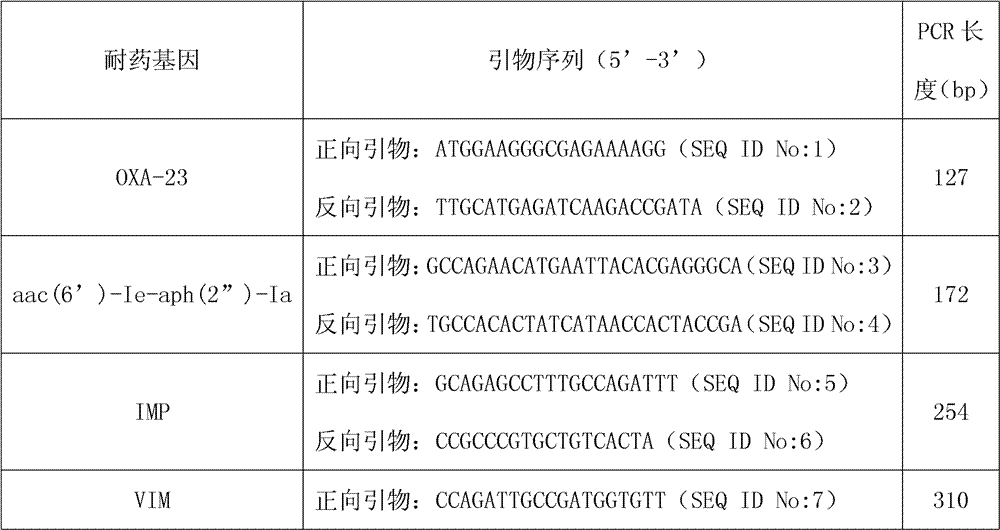
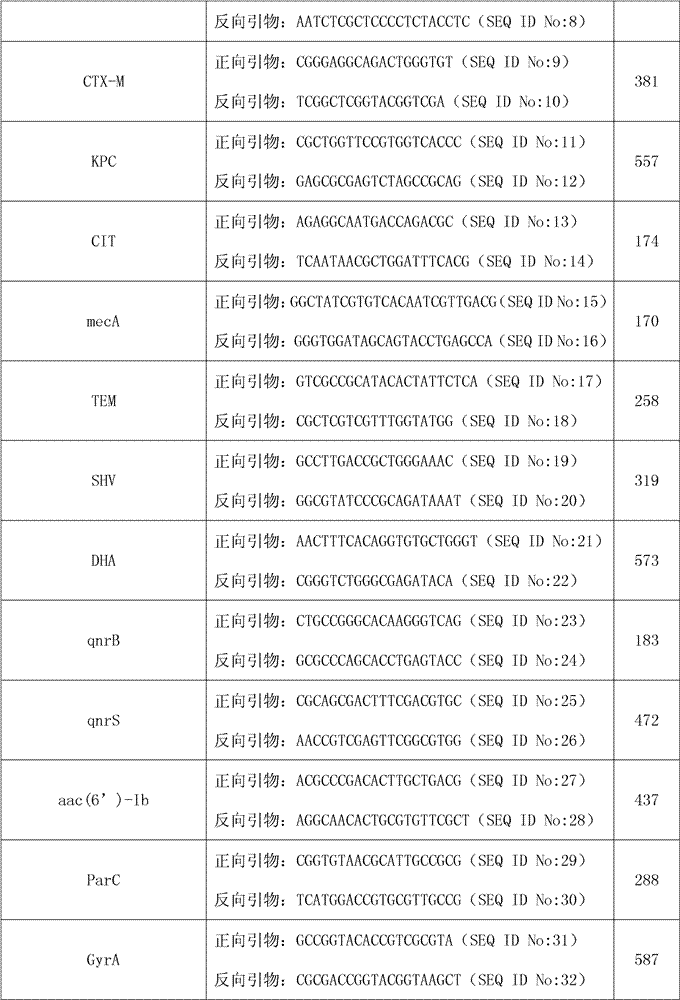
[0031] The drug resistance genes have no bacterial species-specific differences, including TEM, SHV, CTX-M, DHA, CIT, IMP, VIM, KPC, OXA-23, qnrB, qnrS, mecA, aac(6')-Ie- aph(2”)-Ia, aac(6')-Ib, GyrA and ParC, among them, the aac(6')-Ib gene is designed with 3 mutation sites, namely T304C, T304A, G535T; GyrA designed 3 mutations The sites are N87, Y87, and L83; ParC designed 4 mutation sites, namely I80, K84, G84, and V84.
[0032] The sequence length of the forward primer and the reve...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com