Method and primer for detecting lactobacilli in food
A technology for lactic acid bacteria and food, which is applied in the detection of edible lactic acid bacteria and the field of primer pairs, can solve the problems of complex components of lactic acid bacteria products, high cost of DNA sequencing, and long time-consuming, etc., to achieve easy promotion and implementation, low false positive rate, and ensure correctness sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Including the extraction of the bacterial genome of lactic acid bacteria in the food of embodiment 1
[0033] There are 24 imported and exported yogurt samples that can be purchased from the market, including 11 imported products and 13 domestic or joint venture brands. Tiangen Bacterial DNA Extraction Kit (Product No. DP302-02) was used to extract the whole genome of bacteria in 24 yogurt products. DNA. That is, take 1ml of yogurt liquid from different samples, centrifuge at 10,000rpm for 1 minute, discard the supernatant, and then extract the precipitate according to the manufacturer's instructions of the kit to obtain the bacterial genome extract. Determine the DNA concentration by measuring the OD260 / 280 ratio of the extract.
Embodiment 2
[0034] The PCR detection of embodiment 2 lactic acid bacteria strains
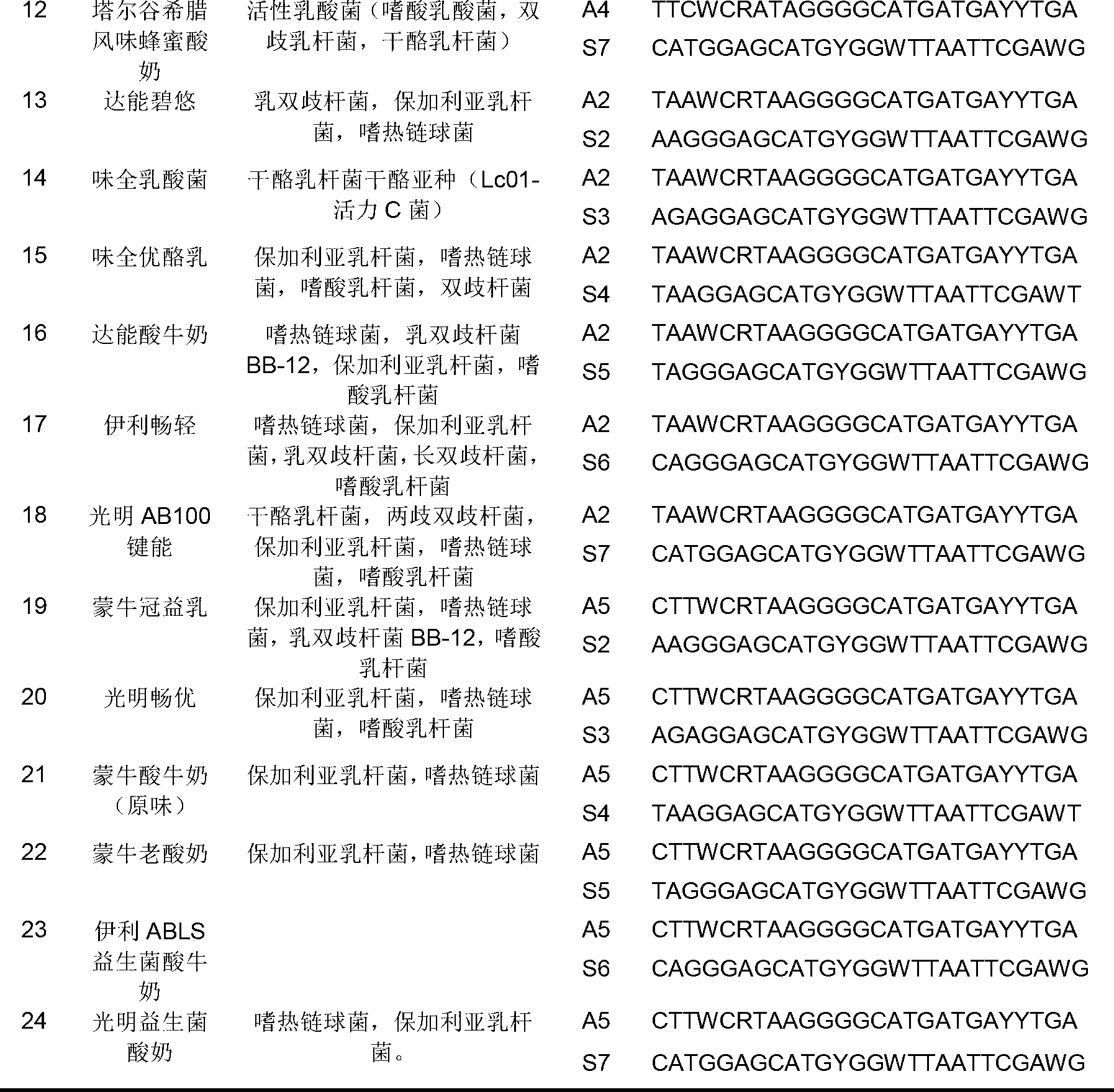
[0035] According to the inventor's research on the safety and sequencing of strains from various manufacturers in the early stage, the merger primer pairs shown in the following table 1 were designed and commissioned by Shanghai Yingjun Biotechnology Co., Ltd. The extracted bacterial genome extracts were subjected to PCR amplification, and the genome extracts of Escherichia coli and Enterobacter sakazakii (also known as Enterobacter sakazakii) were used as controls.
[0036] Table 1
[0037]
[0038]
[0039] Note: In Table 1, W represents equimolar amounts of A and T, Y represents equimolar amounts of T and C, R represents equimolar amounts of A and G; the sequence of primer A2 is shown in SEQ ID NO.1, primer The sequence of A3 is shown in SEQ ID NO.2, the sequence of primer A4 is shown in SEQ ID NO.3, the sequence of primer A5 is shown in SEQ ID NO.4, and the sequence of primer S2 is shown in SEQ ID...
Embodiment 3
[0049] Embodiment 3PCR detects the sequencing verification of the result of lactic acid bacteria strain
[0050] Each batch of the above-mentioned 24 products whose strains were stated to be stable by the manufacturer was tested as in Example 2, 99.2% showed positive results, and 0.8% showed negative results. For further verification, the PCR amplification products used in the positive and negative results were sampled separately for conventional 16S rRNA gene sequencing (using Illumina genome analyzer using 100bppaired-end mode for high-throughput sequencing), and there were no positive results. There was 1 false positive result and 25% of negative results were false negative results. Due to the low false positive rate of the method of the present invention, more than 99% of the workload requiring sequencing can be eliminated even if the sequencing is superimposed.
[0051]
[0052]
[0053]
[0054]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com