Method for quantitatively detecting protein acetylation level
A quantitative detection and acetylation technology, applied in biological testing, material inspection products, fluorescence/phosphorescence, etc., can solve the problems of lack of rapid and non-radioactive quantitative detection of protein acetylation level, difficult antibodies, and difficulties in acetylating proteins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Preparation of protein chip
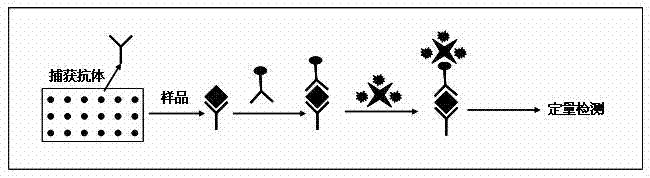
[0028] The bovine serum albumin antibody was spotted on the three-dimensional gel chip using the chip spotting instrument of Beijing Boao Company. The concentration of protein antibody is 200μg / ml, and the instrument maintains 50% humidity. Each chip has a total of 12 matrices. The number of sample points in each matrix is 6 rows and 8 columns, a total of 48 points. Each point is sampled once, and the volume of the antibody is about 1nl. 4°C refrigerator for later use.
Embodiment 2
[0029] Example 2 Detection of Chip Specificity Using Different Kinds of Proteins
[0030] (1) Use bovine serum albumin PBS solution with a mass-volume concentration of 3 μg / ml, acetylated bovine serum albumin PBS solution with a mass-volume concentration of 3 μg / ml, and acetylated horse serum with a mass-volume concentration of 3 μg / ml Myoglobin PBS solution, the horse myoglobin PBS solution that mass-volume concentration is 3 μ g / ml, the goat serum PBS solution that volume percentage concentration is 5%, as the sample of next step;
[0031] (2) The protein chip prepared in Example 1 was blocked with 20 μl of goat serum PBS-T solution with a concentration of 5% by volume for 1.5 h, incubated with 20 μl of the sample for 1.5 h, washed 3 times with TBST solution, and dried; Incubate with 20 μl of biotin-labeled detection antibody with a mass-volume concentration of 2 μg / ml PBS solution for 1.5 h, wash with TBST three times, and dry; Streptomycin PBS solution was incubated for 1...
Embodiment 3
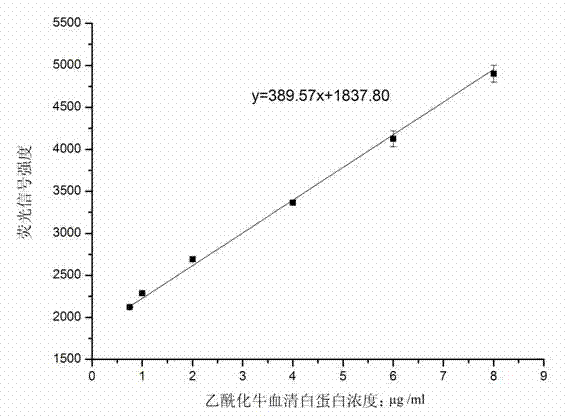
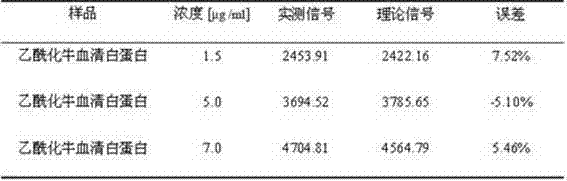
[0037] Example 3 Detect the linear range of acetylated bovine serum albumin in the serum of patients with liver cancer, and draw a standard curve
[0038](1) Dilute with phosphate buffer solution to get 10 times diluted liver cancer patient serum as sample diluent, dilute acetylated bovine serum albumin to obtain standard acetylated bovine serum albumin solutions with different concentrations, the concentrations are 0.25 μg / ml, 0.5μg / ml, 0.75μg / ml, 1μg / ml, 2μg / ml, 4μg / ml, 6μg / ml, 8μg / ml, as the next sample;
[0039] (2) The protein chip prepared in Example 1 was blocked with 20 μl of goat serum PBS-T solution with a concentration of 5% by volume for 1.5 h, then incubated with 20 μl of the sample for 1.5 h, washed 3 times with TBST, and dried; 20 μl mass-volume concentration of 2 μg / ml biotin-labeled detection antibody PBS solution was incubated for 1.5 h, washed 3 times with TBST, and dried; 20 μl mass-volume concentration was 10 μg / ml fluorescence-labeled streptomycin PBS sol...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com