Method for biologically preparing (S)-4-chloro-3-hydroxy butyric acid ethyl ester with recombinant escherichia coli expressed ketoreductase
A technology of recombinant Escherichia coli and ethyl hydroxybutyrate, applied in the biological field, can solve the problems of low optical purity of products and low conversion rate of substrates, achieve high optical purity and reduce production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Acquisition of ketoreductase gene
[0024] according to Candida magnoliae Amino acid sequence of ketoreductase, using E.coli The codon preference was optimized to obtain a new gene sequence (Genbank: KC426948), which was sent to Shanghai Shenggong for synthesis, and NcoI and BamHI restriction sites were added to both ends of the gene. NcoI restriction site: CCATGG, BamHI restriction site: GGATCC.
Embodiment 2
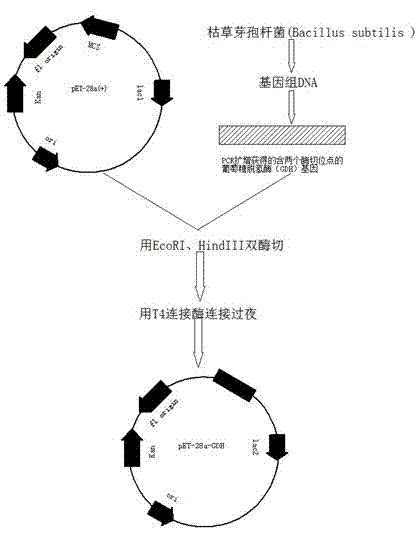
[0025] Example 2 Acquisition of Glucose Dehydrogenase Gene
[0026] Bacillus subtilis ( Bacillus subtilis ) LB liquid medium: tryptone 1%, yeast extract 0.5%, NaCl 1%, pH 7.0.
[0027] Bacillus subtilis was inoculated into 5mL LB liquid medium at 37°C and cultivated to the logarithmic growth phase, and the genome was extracted using a genomic DNA extraction kit (product of TaKaRa).
[0028] The primers used to construct the expression vector are equipped with restriction sites, and the primer sequence is as follows:
[0029] GDH-F1: 5'-CGAATTCATGTATCCGGATTTAAAAGG-3' EcoRI restriction site
[0030] GDH-R1: 5'-GAAGCTTTTATGTTTAACCGCGGCCTG -3' HindIII restriction site
[0031] The primers were synthesized by Shanghai Biological Engineering Co., Ltd.
[0032] Gene PCR (50ul reaction system) conditions are as follows:
[0033] Pre-denaturation at 94°C for 5 minutes; denaturation at 94°C for 45s, annealing at 52°C for 45s, extension at 68°C for 1 min, 30 cycles; extension at 68°C for 10 minute...
Embodiment 3
[0034] Example 3 Expression of ketoreductase gene
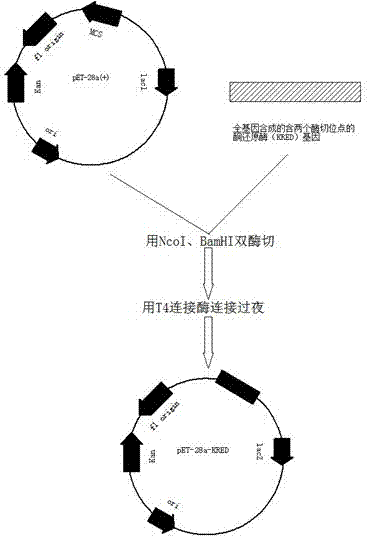
[0035] Use EcoRI and HindIII to digest pET-28a (pET-28a purchased from Novagen (Merck China)) and the synthesized target gene containing two restriction sites, respectively, and recover the double digested target fragment and expression vector. , Connect the double digested expression vector pET-28a and the target gene with T4 ligase overnight, add 10μL of the ligation product pET-28a-ketoreductase to 100μL of BL21(DE3) competent cells, and place on ice 30min, heat shock at 42°C for 90 s. Place on ice for 2 minutes. Add pre-warmed 0.45mL medium. Incubate at 200rpm 37°C for 1h. Add 200μL of bacterial solution to LB plates containing 50μg / mL of kanamycin and chloramphenicol, and incubate overnight at 37°C for 12-16h. Build the map see figure 1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com