Detection method of substance with specific action with nucleic acid beacon
A specificity and beacon technology, applied in the direction of material excitation analysis, fluorescence/phosphorescence, etc., can solve the problems of long preparation cycle, low cost, and reduced material biocompatibility, and achieve simplified preparation difficulty, low-cost detection, biological Effect of Compatibility Improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
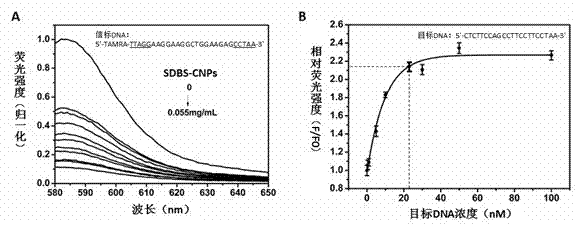
[0027] Take 6 μL of 3 μM tetramethylrhodamine-single-stranded DNA (TAMRA-ssDNA) Tris-HCl (10 mM, pH 7.4, containing 0.15 M NaCl, the same below) solution, add different volumes of 0.1 mg / mL Sodium dodecylbenzenesulfonate stabilized carbon nanoparticle solution (SDBS-CNPs)), add Tris-HCl to make the volume to 600 μL, so that the SDBS-CNPs concentration is 0, 0.01, 0.015, 0.02, 0.025, 0.03, 0.035, 0.04, 0.045, 0.05, 0.055 mg / mL, after incubation at 37°C for 1 h, the down-conversion fluorescence was detected. Prepare a series of Tris-HCl solutions containing different concentrations of target DNA (0, 0.5, 1, 5, 10, 30, 50 and 100 nM), incubate at 42°C for 2 h, then add SDBS-CNPs solution to make the concentration 0.055 mg / mL. Incubate at 37°C for 40 min, detect the down-converted fluorescence, and plot the concentration of target DNA and the ratio of the fluorescence intensity of the sample to the fluorescence intensity of the sample without target DNA. For samples of unknown ...
Embodiment 2
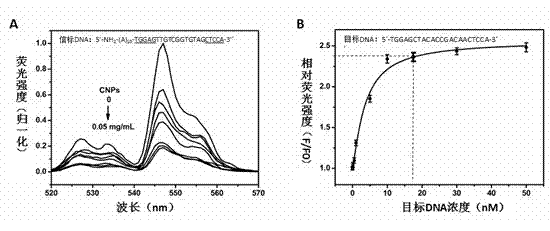
[0030] (1) Preparation of up-converting fluorescent nanoparticles (UCPs): Take 2 ml of 0.25 mol / L rare earth nitrate solution (the molar ratio of rare earth ions is yttrium ion: ytterbium ion: erbium ion = 80:18:2, add Add 18 ml of absolute ethanol, then add 8 ml of an aqueous solution containing 900 mg of polyacrylic acid, and stir for 10 min; add 8 ml of an aqueous solution containing 0.210 g of sodium fluoride to the above mixed solution, continue stirring for 20 min, and place it under high pressure for reaction In the kettle, under stirring conditions, hydrothermal reaction was carried out at 200 °C for 10 h; stop heating and keep stirring to cool to room temperature, centrifuge to separate the solid product, wash with absolute ethanol and ultrapure water three times, and vacuum dry at room temperature for 12 h , to obtain up-converting fluorescent nanoparticles with carboxyl groups on the surface.
[0031] (2) Surface labeling of UCPs: Dissolve 5 mg of the upconvertin...
Embodiment 3
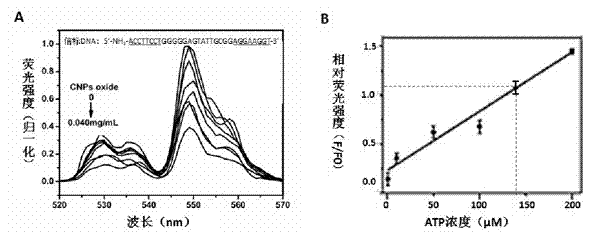
[0036] Take 30 μL of 0.6 mg / mL UCPs Tris-HCl solution labeled with beacon DNA (the synthesis and labeling methods of UCPs are the same as in Example 2), add different amounts of carbon dioxide nanoparticles (CNPs oxide) aqueous solution, and constant volume to 600 μL. After incubation at 30°C for 90 min, upconversion fluorescence was detected using 980 nm excitation light. Add different amounts of adenosine triphosphate (ATP) to the Tris-HCl solution containing 0.03 mg / mL beacon DNA-UCPs and 0.04 mg / mL CNPs oxide, incubate at 30°C for 90 min, and measure the upconversion fluorescence with 980 nm excitation light. Calculate the ratio of the fluorescence intensity F of the sample added with ATP to the fluorescence intensity F0 of the sample without ATP, and plot the ratio of ATP concentration to F / F0 to obtain a standard curve. For samples with unknown concentrations, add beacon DNA-UCPs solution and CNPs oxide aqueous solution and add Tris-HCl to make up to 600 μL, so that...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com