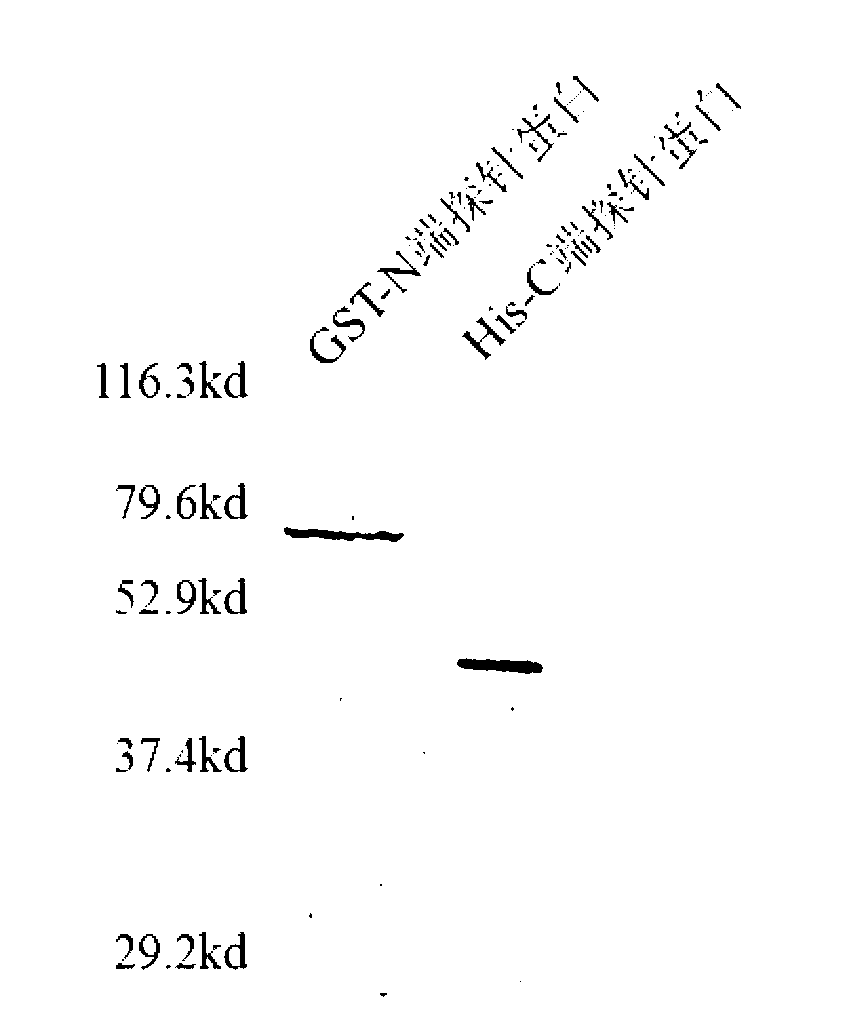Metastasis suppressor1 protein dimerization fluorescent probes and application thereof
A technique for metastatic tumors and fluorescent probes, applied in the field of fluorescent probe complementary proteins, to achieve the effects of simple processing, few steps and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
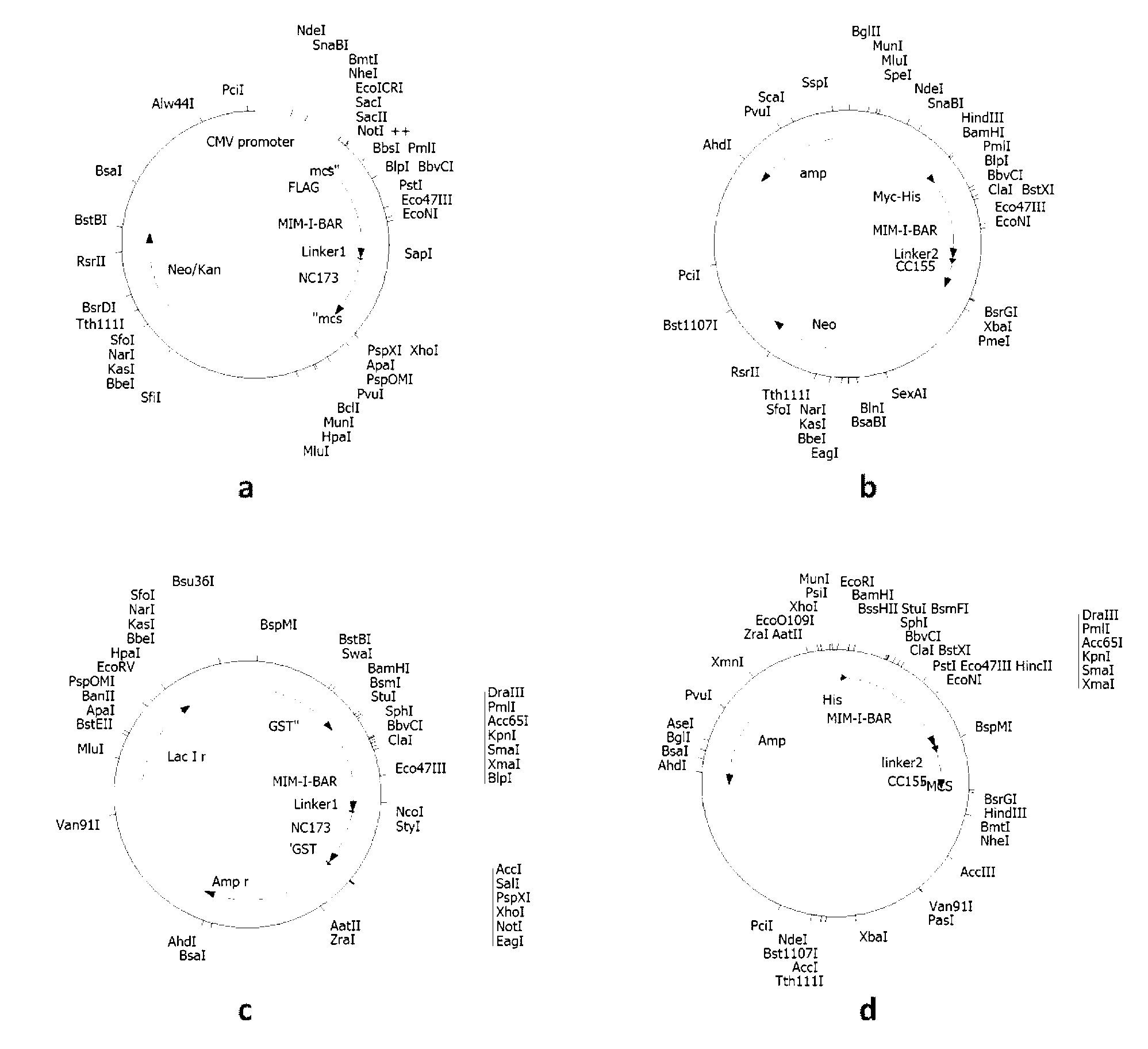
[0026] Example 1 Construction of plasmid DNA comprising polynucleotides represented by SEQ ID No: 1 and SEQ ID No: 3 respectively introducing Flag, Myc, GST and His tags.
[0027] (1) Construction of the N-terminal protein-encoding gene carrier of the fluorescent probe represented by Flag-labeled SEQ ID No: 2, that is, the plasmid (also known as pFlag-Mtss1-N) containing the polynucleotide represented by SEQ ID No: 1
[0028] The insert fragment was prepared using the overlap extension PCR method as follows: the DNA fragment Mtss1 (GenBankSEQ ID No: 211401) was used as a template, and the polynucleotides of 5'-CAGTTAGGATCCGAGGCTGTGTGATCGAG-3' and 5'-CGTAGCGATAGATCGGGGAGAAGAAGG-3' sequences were used as primers, Perform PCR amplification. The amplification reaction conditions are: 50μl reaction system, using PfuUltra TM II fusion HS DNA polymerase and its matching 10x buffer, 250 μM each of dNTP, 20 ng / μl template DNA, and 0.5 μM each primer. 30 cycles were performed on the M...
Embodiment 2
[0042] Example 2 Transfection and expression of fluorescent probe complementary proteins represented by SEQ ID No: 2 and 4 with Flag, Myc, GST, His tags, and corresponding polynucleotide recombinant plasmids in eukaryotic / prokaryotic cells.
[0043] Human embryonic kidney cell 293T cells were cultured using DMEM high-glucose medium containing 10% calf serum BCS, adding penicillin at a final concentration of 100 units / ml and streptomycin at a final concentration of 100 μg / ml. Before DNA transfection, 1.5x10 6 Cells were seeded in 100mm Petri dishes using normal medium without antibiotics at 37°C and 5% CO 2 conditions overnight. The transfection mixture was then prepared: 5 μg plasmid pFlag-Mtss1-N, 5 μg plasmid pMyc-Mtss1-C, 300 μl DMEM, 60 μl SuperFect. After the transfection mixture was incubated at room temperature for 5-10 minutes, add 3ml of normal culture medium and mix well to replace the cell culture medium and culture the cells for 2-3 hours, then replace it with no...
Embodiment 3
[0046] Example 3 The corresponding recombinant plasmid expression vector comprising the polynucleotide represented by SEQ ID No: 1 and 3, and the detection of the expression product.
[0047] After eukaryotic or prokaryotic cells are transfected with recombinant plasmid expression vectors, the detection and evaluation methods of expression products mainly include Western Blot detection method, Coomassie brilliant blue detection method and immunofluorescence detection method.
[0048] Western Blot detection method is as follows. Chromatography of cell lysates or purified protein samples by 10% SDS-polyacrylamide gel electrophoresis. After the electrophoresis, wet electrotransfer the gel to the nitrocellulose membrane at 350mA current at 4°C for 1h, incubate overnight at 4°C with the 1:500-1:2000 dilution of 5% skimmed milk of the corresponding antibody, and buffer with TBST. After washing with horseradish peroxidase-labeled secondary antibody IgG1:200~1:1000, incubate at room te...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com