Gallic acid base light curing active monomer, preparation method and application thereof
A technology of gallic acid and active monomers, applied in the field of bio-based polymer materials, can solve the problems of difficult to obtain performance photocurable coatings, flexible aliphatic chain length, low reactivity, etc. simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
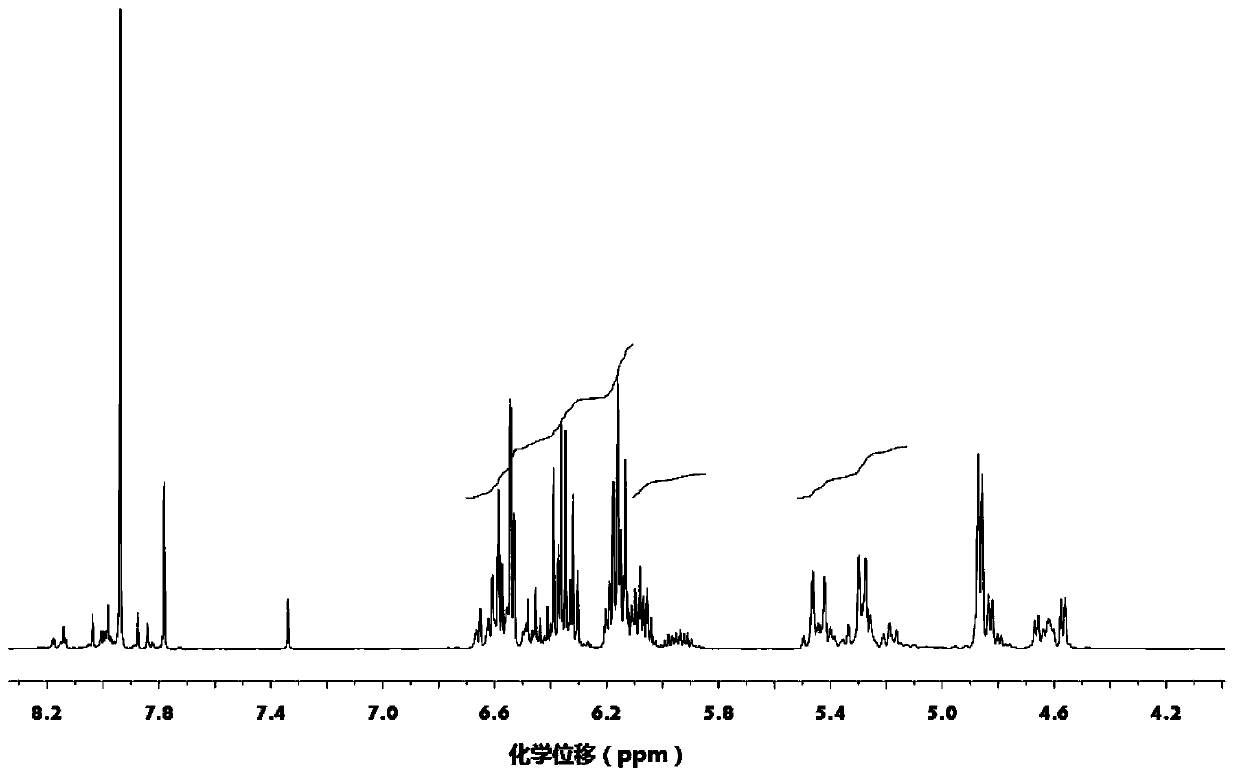
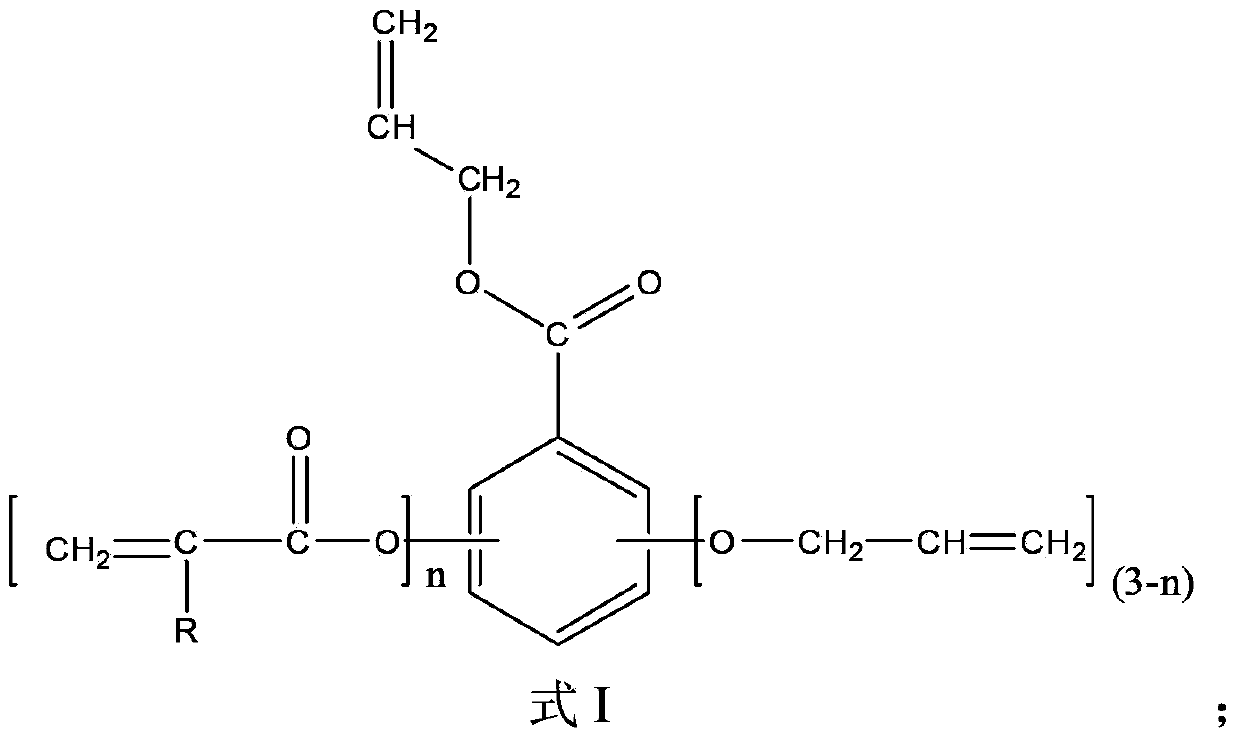
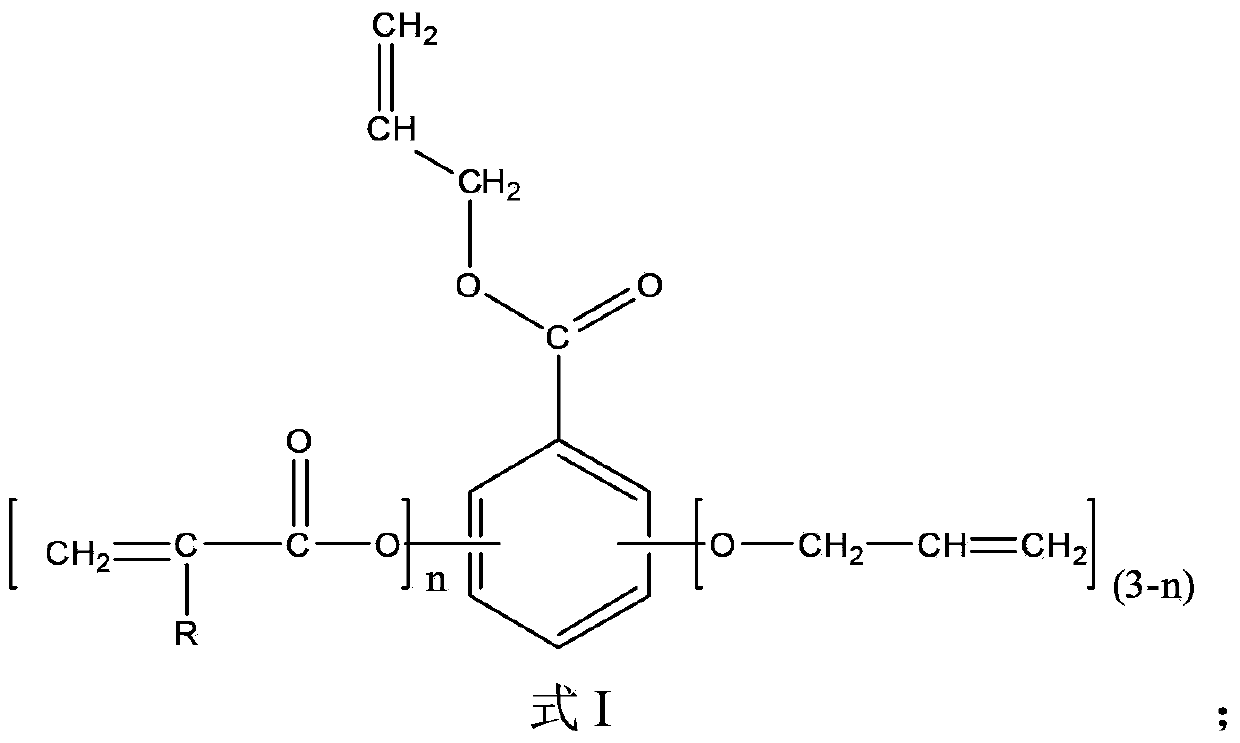
[0031] Mix 100 parts by mass of gallic acid, 1500 parts by mass of acetone, 180 parts by mass of allyl chloride and 200 parts by mass of calcium oxide, react at 50°C for 48 hours, then add 250 parts by mass of acryloyl chloride, Continue to react for 48 hours, filter, wash with water, obtain final product after decompression distillation removes solvent, water etc., the product 1 HNMR spectrum as figure 1 as shown, figure 1 The appearance of 5.21-5.49ppm and 5.84-6.13ppm represents the H on the allyl double bond, and the peak at 6.11-6.65ppm represents the H on the acryloyl double bond, plus other peaks are related to the gallic acid-based photocurable active unit The H proton displacement of the body is consistent, proving that the obtained product is a gallic acid-based photocurable active monomer of the formula I structure; from the peak area corresponding to the H on the allyl double bond and the H on the acryloyl double bond, it can be calculated out n=1.7
[0032] ...
Embodiment 2
[0036] 100 mass parts of gallic acid, 1000 mass parts of methyl ethyl ketone, 400 mass parts of N,N'-dimethylformamide, 100 mass parts of allyl bromide and 200 mass parts of triethylamine were mixed at 20 ℃ for 12 hours, then add 400 parts by mass of acryloyl chloride, react at 40 ℃ for 24 hours, filter, wash with water, and distill under reduced pressure to remove solvent, water, etc. to obtain the final product. 1 The appearance of 5.21-5.49ppm and 5.84-6.13ppm in the HNMR spectrum represents the H on the allyl double bond, and the peak at 6.11-6.65ppm represents the H on the acryloyl double bond, plus other peaks and gallic acid group light The H proton displacement of the curing active monomer is consistent, proving that the obtained product is a gallic acid-based photocuring active monomer of the formula I structure; the peak area corresponding to the H on the allyl double bond and the H on the acryloyl double bond , n=3 can be calculated.
[0037]
[0038] Wherein, R...
Embodiment 3
[0040] Mix 100 parts by mass of gallic acid, 1000 parts by mass of dioxane, 270 parts by mass of allyl chloride and 60 parts by mass of sodium hydroxide, react at 60°C for 24 hours, and then add 500 parts by mass of dimethyl sulfoxide, 100 parts by mass of acryloyl chloride and 40 parts by mass of sodium hydroxide were mixed, reacted at 50°C for 12 hours, filtered, washed with water, and evaporated under reduced pressure to remove solvent and water to obtain the final product. 1The appearance of 5.21-5.49ppm and 5.84-6.13ppm in the HNMR spectrum represents the H on the allyl double bond, and the peak at 6.11-6.65ppm represents the H on the acryloyl double bond, plus other peaks and gallic acid group light The H proton displacement of the curing active monomer is consistent, proving that the obtained product is a gallic acid-based photocuring active monomer of the formula I structure; the peak area corresponding to the H on the allyl double bond and the H on the acryloyl double ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Glass transition temperature | aaaaa | aaaaa |
| Tensile strength | aaaaa | aaaaa |
| Glass transition temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com