Gluconobacter oxydans gene engineering bacteria for producing 2-KLG and its application
A technology of gluconic acid bacteria and genetically engineered bacteria, applied in the field of genetic engineering, achieving good application prospects, simple construction methods, and simplified production processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] The construction of embodiment 1 expression vector
[0025] The sdh and sndh gene sequences annotated in the whole genome sequencing results of K. vulgare WSH-001 in our laboratory were connected with 10 kinds of connecting peptides (GGGGS, GGGGSGGGGS, GGGGSGGGGSGGGGS, PTPTP, PTPTPTPTP, PTPTPTPTPTPTPTP, EAAAK, EAAAKEAAAK, EAAAKEAAAKEAAAK and SSSNNNNNNNNNNNN) primers were amplified, and sequenced by connecting the cloning vector pMD19-T after PCR fusion. At the same time, the strong promoter tufB derived from G.oxydans was amplified and connected to the cloning vector pMD19-T for sequencing to obtain the correct transformation Afterwards, double digestion was performed and connected to the E.coli-G.oxydans shuttle plasmid vector pGUC1 to construct 10 fusion expression vectors pGUC-tufB-sdh-GS-sndh, pGUC-tufB-sdh-GS 2 -sndh, pGUC-tufB-sdh-GS 3 -sndh, pGUC-tufB-sdh-PT-sndh, pGUC-tufB-sdh-PT 4 -sndh, pGUC-tufB-sdh-PT 7 -sndh, pGUC-tufB-sdh-EAK-sndh, pGUC-tufB-sdh-EAK 2 ...
Embodiment 2
[0026] The construction of embodiment 2G.oxydans engineering bacteria
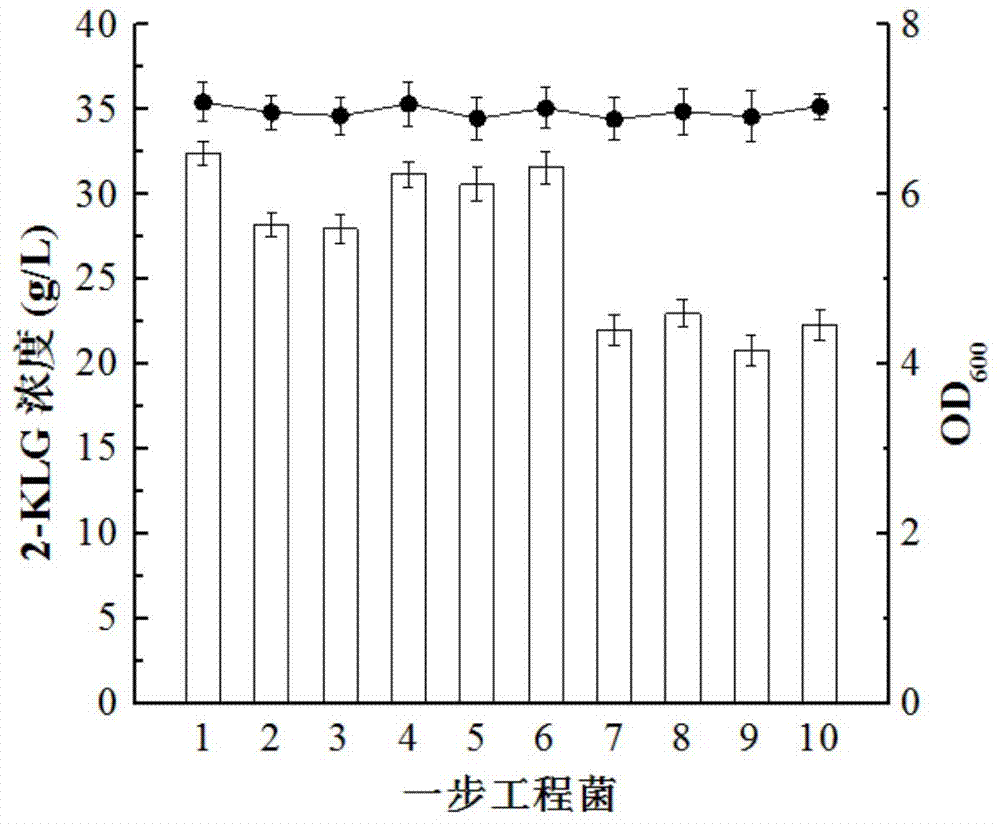
[0027]The constructed expression vector was transformed into E.coli JM109, spread on the LB medium containing ampicillin (yeast extract 5g / L, peptone 10g / L, NaCl10g / L, solid medium plus 20g / L agar, 121 ℃ sterilized for 15min), pick the transformant on the transformed plate for PCR verification, and a band of about 3027bp appears, which proves that it has been successfully transformed into E.coli JM109, and then transferred to G.oxydans WSH by the method of three-parent hybridization In -003, 10 strains of G.oxydans engineering bacteria were obtained.
Embodiment 3
[0028] Embodiment 3 fermentation produces 2-KLG
[0029] Seed medium (g / L): sorbitol 15, yeast powder 1, pH 4.8-5.1, agar 20 (solid medium), sterilized at 121°C for 15 minutes, final concentration of ampicillin 100 μg / mL.
[0030] Fermentation medium (g / L): sorbitol 15, yeast extract 1.2, calcium chloride 0.2, initial pH 5.1-5.4, sterilized at 121°C for 15 minutes, final concentration of ampicillin 100 μg / mL.
[0031] Culture conditions: Scrape a few rings of bacteria from the solid plate and inoculate them into a 500mL double-thorn shaker flask filled with 50mL of liquid medium (added with a final concentration of 75μg / mL ampicillin), and culture in a rotary shaker at 30°C at 200r / min When reaching the logarithmic growth phase (about 30h), transfer it to a fresh medium with a final concentration of 75 μg / mL ampicillin according to 15% (v / v) inoculation amount, and then culture until the logarithmic growth phase. ) inoculum amount was transferred to the fermentation medium, 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com