Fluorescent quantitative PCR primers and probes for porcine epidemic diarrhea viruses
A swine epidemic diarrhea, fluorescence quantitative technology, applied in biochemical equipment and methods, microbial determination/inspection, microorganisms, etc., can solve the problems of pig industry losses, lack of fluorescence quantitative differential diagnosis methods, etc., to achieve primer specificity and high sensitivity, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] (1) RNA extraction of the sample to be tested: 200 μL cell culture medium (infected with the self-isolated strain CHGD-01, which has been identified as a mutated PEDV, and other mutated strains can also be used), 200 μL of cell culture medium (infected with the self-isolated strain strain ShQT, which has been identified as classic PEDV, and other classic strains can also be used), 100 mg of intestinal disease material and 100 mg of feces (the disease material and feces are unknown whether the virus is infected), and DMEM (cell culture medium, without PEDV, as a negative control) as the sample to be tested, the following operations were performed respectively:
[0035] Add 1mL of PBS buffer for thorough grinding, repeat freeze-thaw at -20°C for 3 times, centrifuge at 8000 rpm for 10 min, take 200 μL of supernatant, add 0.2mL of chloroform, shake and mix for 15 seconds, and then store at room temperature (15°C-30°C) After standing for 2-3 minutes, centrifuge at 12000g (2°...
Embodiment 2
[0044] Example 2 specificity test
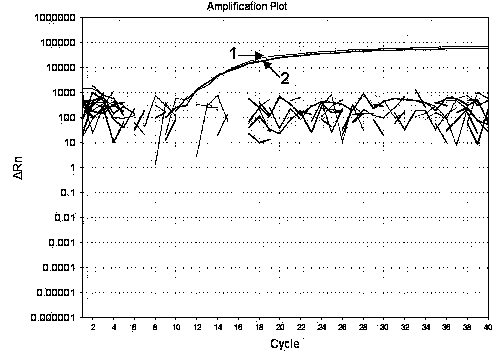
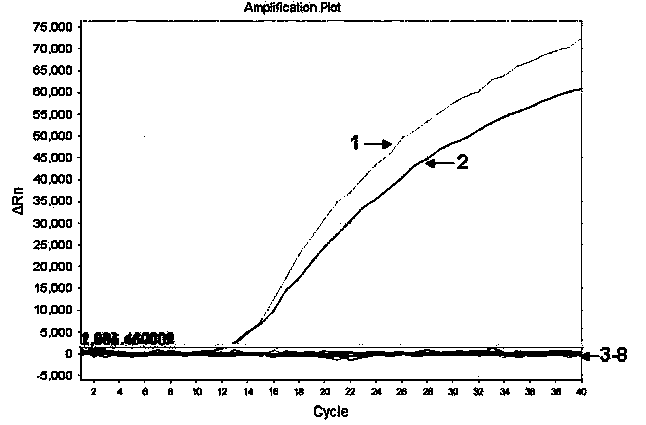
[0045] Porcine transmissible gastroenteritis virus, porcine rotavirus, porcine reproductive and respiratory disorder syndrome The culture fluid of porcine circovirus, porcine circovirus, classical swine fever virus, and porcine pseudorabies virus is specifically detected, and the method, primers and probes of Example 1 are used. As a result, there is no amplification curve except the positive control, and the positive disease material After PCR amplification, there are amplification curves in HEX (strain ShQT) and FAM (strain CHGD-01) groups respectively (see figure 2 ).
Embodiment 3
[0046] Embodiment 3 Sensitivity test
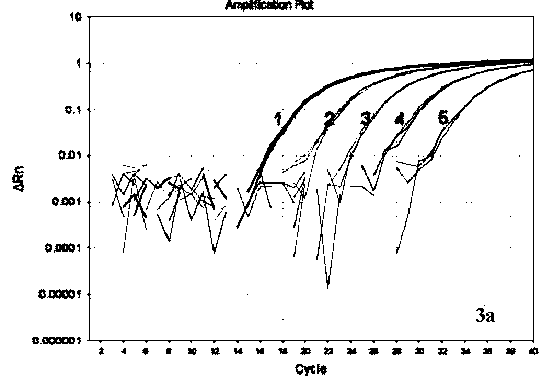
[0047] Standard positive plasmids (pMD-S1 and pMD-S2) containing the S gene, 191bp amplified by the PCR involved in this patent (use the cDNA extracted from the mutant strain ShQT as a template, and use SEQ ID NO: 1~2 as primers for PCR , the amplified product sequence is shown in SEQ ID NO.5) and 179bp (with the cDNA extracted from the classical strain CHGD-01 as a template, and SEQ ID NO: 1~2 are primers for PCR, the amplified product sequence is shown in SEQ ID NO .6) was connected to the cloning vector pMD18-T (Dalian Bao Biological Engineering Co., Ltd.), and its concentration was determined to be 3.94×10 10 copies / μL and 2.99×10 10 copies / μL.
[0048] The positive plasmids pMD-S1 and pMD-S2 were diluted 10 times with the prepared sterile double distilled water as the template, and 2 μL was taken for each dilution as the template, and the detection was carried out according to the method in Example 1, and the amplification curv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com