Spinal muscular atrophy-related gene mutation detection method, related detection probe composition and detection kit as well as related application
A spinal muscular atrophy and detection kit technology, applied in the field of disease-related gene mutation detection and spinal muscular atrophy-related gene mutation detection, can solve problems such as easy contamination, avoid sample contamination, rapid detection, and clever design Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: DNA preparation
[0025] 1) Preparation of normal human genomic DNA for testing
[0026] Take normal human blood filter paper, extract and prepare by Chlex100, and adjust the concentration of normal human genomic DNA to 10ng / μl.
[0027] 2) SMN1 normal genotype standard preparation
[0028] According to the sequences of SMN1 and internal reference gene β-actin, SMN1EXON7 (SEQ ID NO:1), SMN1EXON8 (SEQ ID NO:2) and β-actin (SEQ ID NO:3) were artificially synthesized. The concentration of the T vector containing the above fragments was adjusted to 1ng / ul as a 10-fold normal genotype template.
[0029] 3) SMN1 mutation deletion genotype standard preparation
[0030]The internal reference gene β-actin (SEQ ID NO:3) was artificially synthesized. The concentration of the T vector containing the above fragments was adjusted to 1ng / ul as a template for the 10-fold mutation deletion genotype.
Embodiment 2
[0031] Embodiment 2: Design and synthesis of SMN1 mutant gene amplification primers and MGB probes
[0032] Synthetic primers were designed according to SMN1EXON7 and SMN1EXON8, the SMN1 gene probe was labeled Fam, and the internal reference gene probe was labeled Vic. Verify that fluorescent quantitative PCR primers specifically amplify target gene fragments, and Taqman MGB site-specific probes report corresponding fluorescent signals. Artificially synthesized gene fragment-specific amplification primers and site-specific Taqman MGB reporter probes. See Table 1 and Table 2 for gene fragment-specific primers and site-specific Taqman MGB probe sequences (SEQ ID NO: 4 to SEQ ID NO: 12).
[0033] Table 1
[0034]
[0035]
[0036] Table 2
[0037]
Embodiment 3
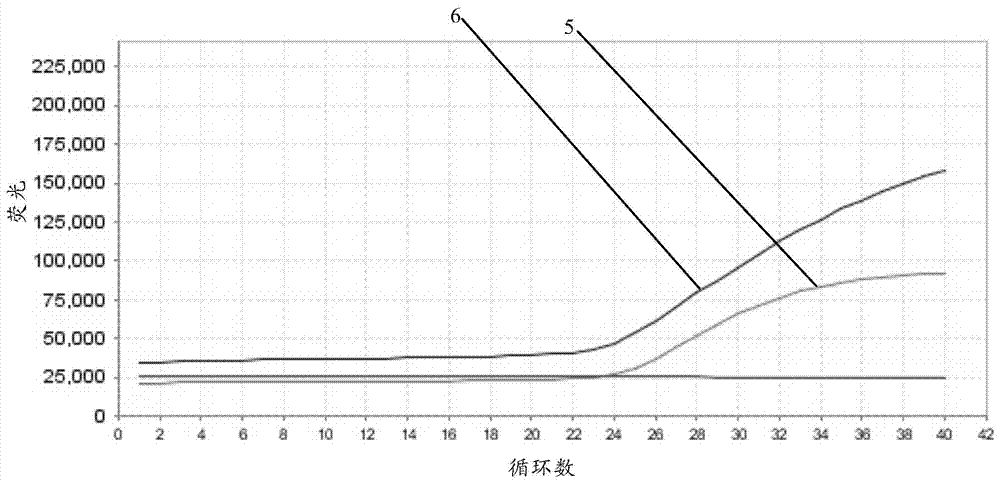
[0038] Example 3: Normal or missing detection of SMN1 Taqman quantitative PCR gene
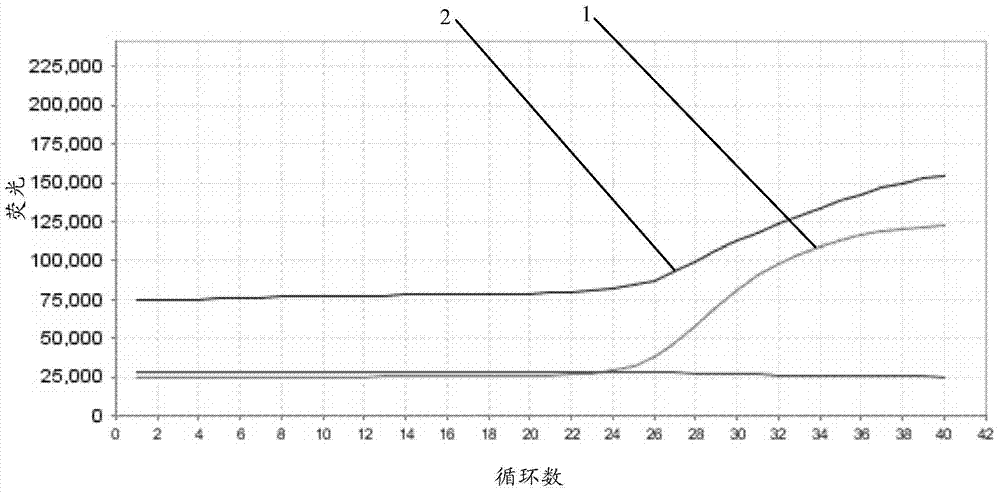
[0039] 1) exon7 gene verification
[0040] Reaction system: 2.5 μl of primer-probe mixture, 12.5 μl of 2-fold Multiplex PCR Mix (QIAGEN N.V., Germany), 2.5 μl of standard template, 5 μl of 5-fold Easy Buffer, ddH 2 O2.5 μl.
[0041] Quantitative PCR instrument: ABI7500 (Life Technologies, USA).
[0042] Reaction conditions: 95°C for 5 minutes; 94°C for 30 seconds—60°C for 30 seconds—72°C for 45 seconds, 40 cycles.
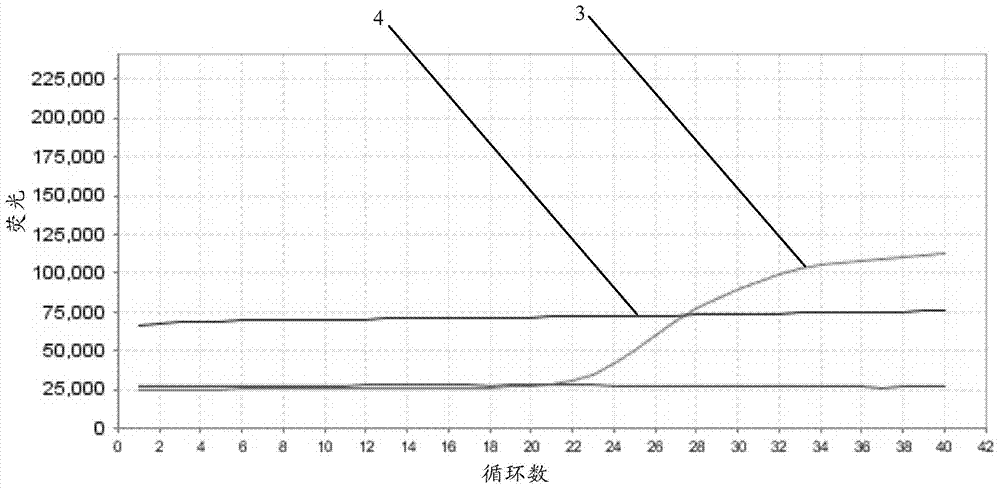
[0043] 2) exon8 gene verification
[0044] Reaction system: 2.5 μl of primer-probe mixture, 12.5 μl of 2-fold Multiplex PCR Mix (QIAGEN N.V., Germany), 2.5 μl of standard template, 5 μl of 5-fold Easy Buffer, ddH 2 O2.5 μl.
[0045] Quantitative PCR instrument: ABI7500 (Life Technologies, USA).
[0046] Reaction conditions: 95°C for 5 minutes; 94°C for 30 seconds—60°C for 30 seconds—72°C for 45 seconds, 40 cycles.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com