Quantitative detection kit of hepatitis A virus
A hepatitis A virus, quantitative detection technology, applied in the determination/inspection of microorganisms, resistance to vector-borne diseases, biochemical equipment and methods, etc. Accuracy and traceability, guaranteed precision and accuracy, good detection specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Fluorescent quantitative PCR primer design and specificity verification
[0033] By consulting the relevant information of hepatitis A virus, SYBR Green real-time PCR primers were designed with Beacon Designer7 software. Select the conserved region encoding the VP1-VP3 capsid protein in the hepatitis A virus genome, and design the upstream and downstream primers of the gene fragment in the conserved region encoding the VP1-VP3 capsid protein in the hepatitis A virus genome. The sequences are SEQ ID NO: 1~SEQ ID NO: 2 (Table 1), the length of the amplicon is 112bp; use the Bio-Rad CFX Manager instrument to perform temperature gradient fluorescent quantitative PCR in the range of 43.0°C-63.0°C, optimize the annealing temperature to 55°C, and determine the primer effect.
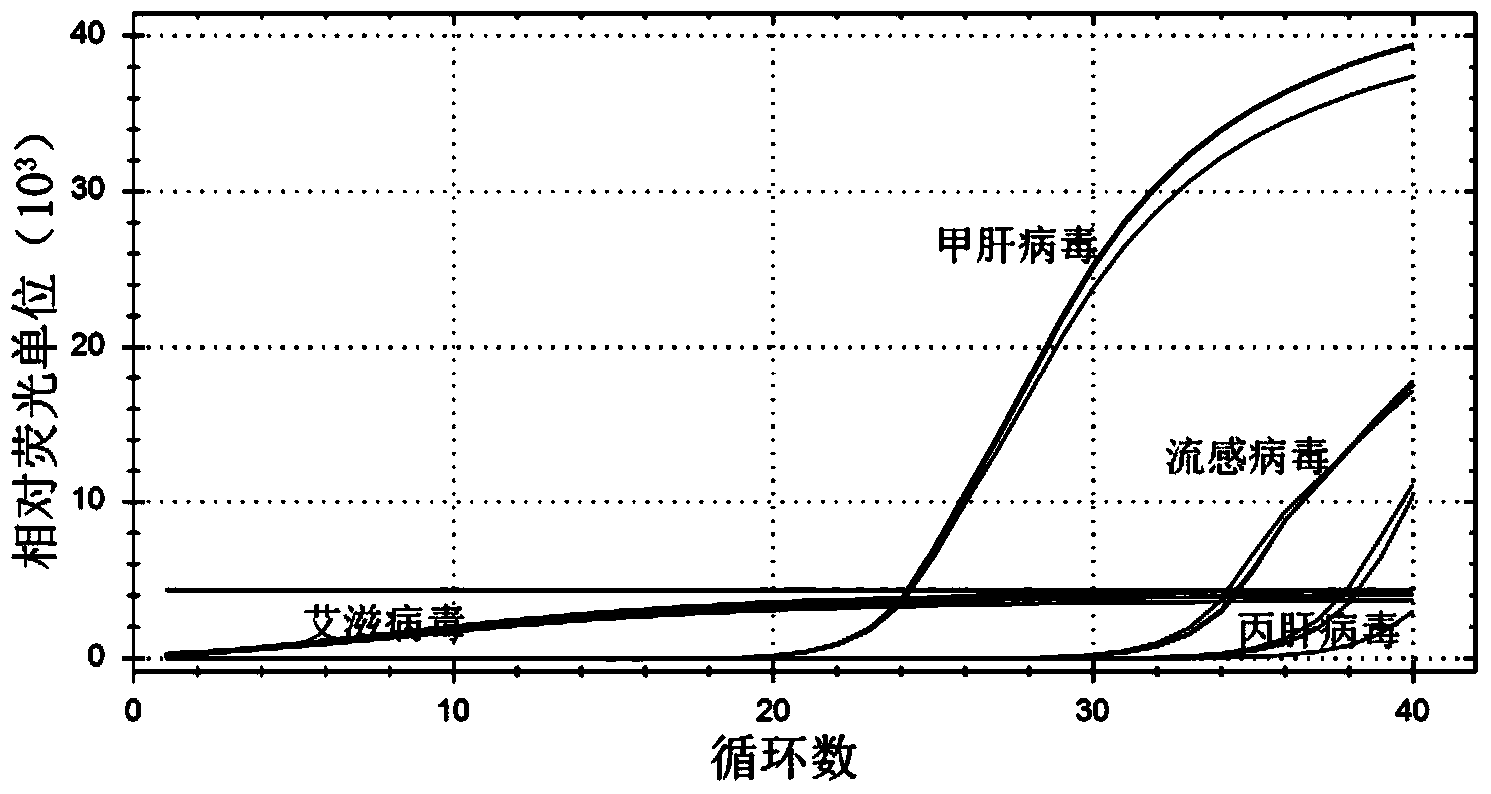
[0034] Using the hepatitis A virus genome cDNA as a template, use non-hepatitis A virus primers such as specific detection primers for influenza virus, hepatitis C virus, and HIV for fluoresce...
Embodiment 2
[0035] Embodiment 2: the drawing of standard curve
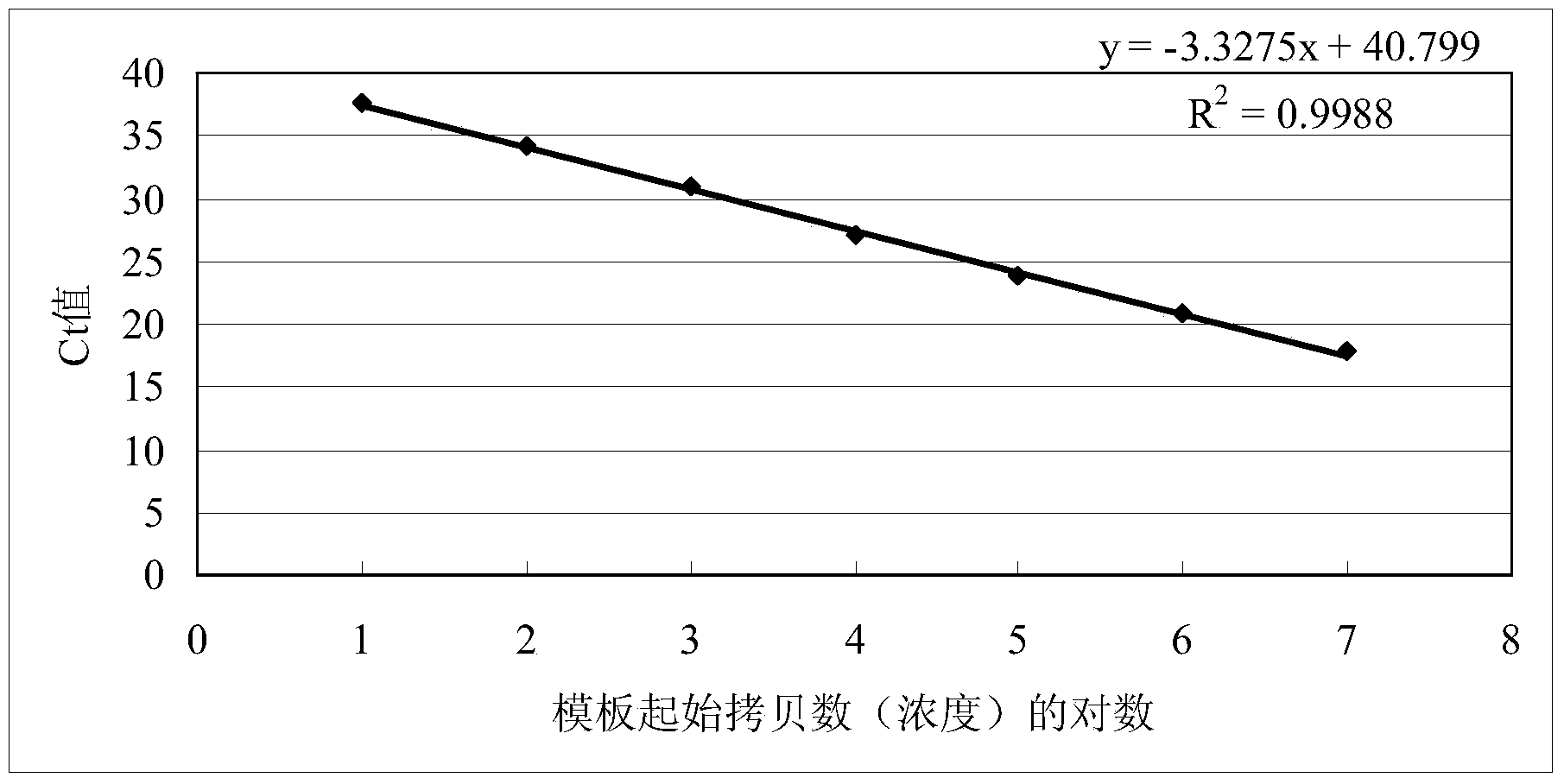
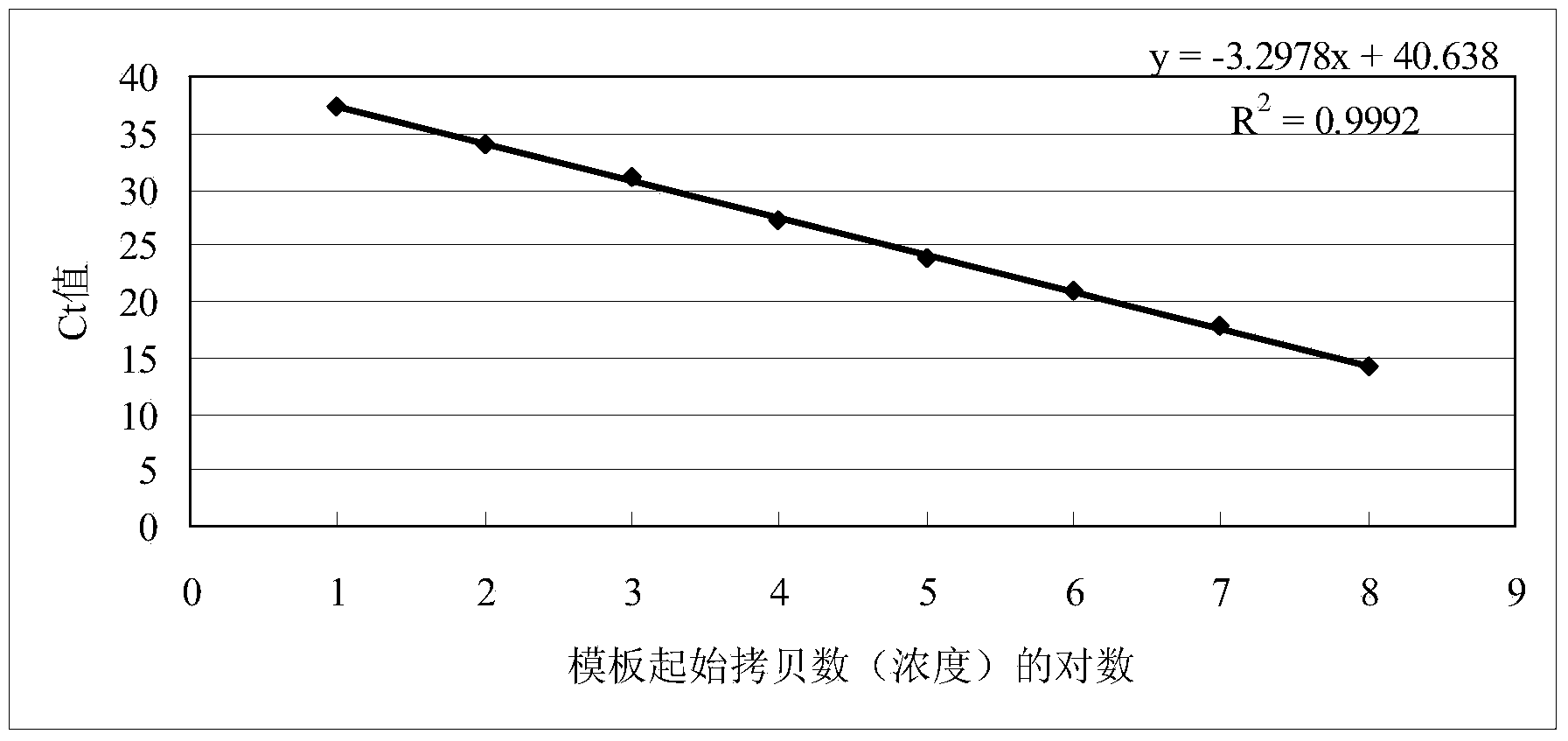
[0036] Dilute the standard with ddH 2 O diluted to 1.0 x 10 7After copy / μL, perform 10-fold dilutions sequentially, use dilutions of different concentrations as templates, and take 10 for the standard curve 1 、10 2 、10 3 、10 4 、10 5 、10 6 、10 7 7 spots for copies / μL. Such as figure 2 Shown, different dilutions of hepatitis A virus standard (10-10 7 copy / μL), the amplification curve is clear and the spacing is uniform. The linear relationship of the standard curve is good Y=-3.3275X+40.799, R 2 =0.9988, the amplification efficiency was 99.77%, and the detection limit was as low as 10 copies / μL.
[0037] Dilute the standard with ddH 2 O diluted to 1.0 x 10 8 After copy / μL, perform 10-fold dilutions sequentially, use dilutions of different concentrations as templates, and take 10 for the standard curve 1 、10 2 、10 3 、10 4 、10 5 、10 6 、10 7 、10 8 8 spots for copies / μL. Such as image 3 Shown, different d...
Embodiment 3
[0038] Example 3: Extracting viral RNA and reverse transcription.
[0039] Extract viral RNA: Add Trizol and chloroform to the EP tube containing hepatitis A virus (the volume ratio of each component added is: virus liquid: Trizol: chloroform = 1:2:1), shake for 30 seconds, and let stand on ice for 5 minutes Finally, centrifuge at 12000r / min at 4°C for 15min; gently absorb the upper aqueous phase, put it in a new EP tube, and discard the rest. Add an equal volume of isopropanol and 1 μL Glyco Blue Coprecip (Invitrogen) to the EP tube containing the aqueous phase liquid, mix well, place in a -20 °C refrigerator for 2 min, centrifuge at 12000 r / min, 4 °C for 10 min; discard the supernatant Wash the precipitate with 200ml of 75% ethanol (made with RNase-free water), centrifuge at 12000r / min, 4°C for 10min; discard the supernatant, and let the precipitated RNA dry naturally at room temperature; dissolve the RNA with 20μL RNase-free water .
[0040] The reverse transcription syst...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com