Establishment method of clostridium difficile exotoxin A carboxyl-terminal sequence codon optimization gene segment and expression vector and application of expression protein thereof
A technology of codon optimization and Clostridium difficile, applied in the direction of anti-bacterial immunoglobulin, biochemical equipment and methods, applications, etc., can solve the problems of codon self-preference difference and low expression efficiency of Escherichia coli
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Codon-optimized Clostridium difficile exotoxin A carboxy-terminal gene sequence design
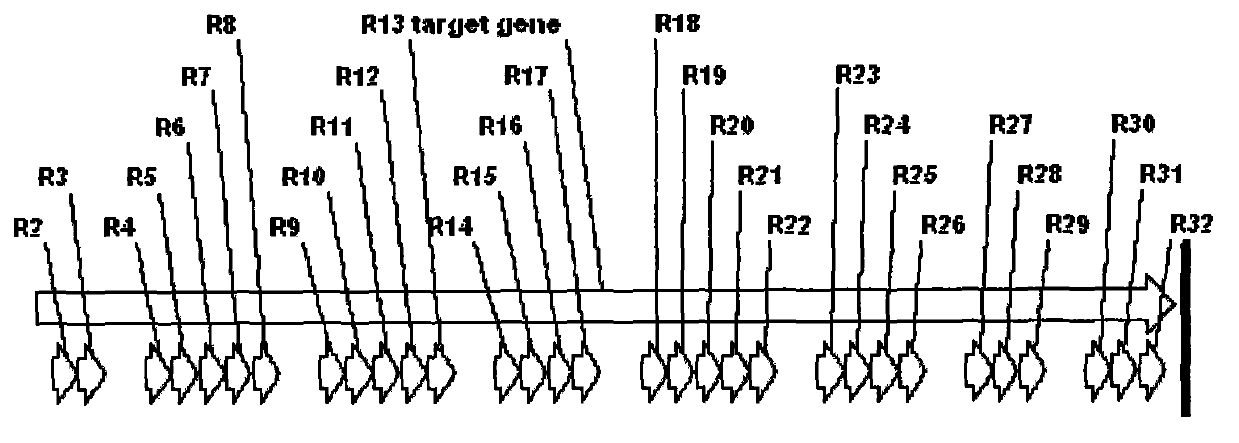
[0042] Select the 2616-base sequence at the carboxy-terminal of the Clostridium difficile exotoxin A gene (encoding 872 amino acids in total), analyze the coding sequence, and find out the repetitive sequence that affects gene synthesis. The schematic diagram of the repetitive region is shown in figure 1 . Simultaneously analyze the sites with different codon usage preferences in this sequence and E. coli, and replace low-frequency codons with high-frequency codons; in addition, on the basis of taking into account the codon preferences of E. The sequence of the synonymous codon was replaced to obtain a new optimized coding gene sequence.
[0043] The method for codon modification includes the following aspects, that is, based on the homology analysis of the amino acid sequence of the repeat region (see figure 2 ), find out the conserved amino acid sequence (conservative...
Embodiment 2
[0051] Example 2 Codon-optimized Clostridium difficile exotoxin A carboxy-terminal gene sequence synthesis
[0052] According to the optimization principle of Example 1 above, the corresponding nucleotide sequence was designed with reference to the codon preference of Escherichia coli, and the carboxy-terminal gene of the C. difficile exotoxin A gene after codon optimization was sent to gene synthesis in two stages. Restriction sites are designed at both ends of each gene for subsequent molecular cloning.
[0053] The synthetic sequence of the first optimized combined sequence SEQ ID NO.29 Fragment 1 (SEQ ID NO.32) is as follows. Introduce the EcoRI digestion recognition sequence "GAATTC" at the end:
[0054] 5'gatatctcat tattctattt tgatcctata gaatttaact tagtaacagg atggcaaact 60
[0055] atcaacggca agaaatatta cttcgatata aatactggag cagctttaac tagttataag 120
[0056] atttattaatg gtaaacactt ttactttaat aatgatggtg tgatgcagct gggagtattt 180
[0057] aaaggacctg atggatttga atatttt...
Embodiment 3
[0110]Example 3 Construction and identification of the first optimized combination prokaryotic expression vector pET3b(+)-TcdA-C-1
[0111] 1. Acquisition of the target gene fragment: Use EcoRV and EcoRI to double digest the subcloning plasmid pUCE-TcdA-C1 (synthesized by Gene Company, containing the first combined optimized fragment 1 sequence), recover the fragment 1 sequence by gel recovery; use EcoRI and XhoI double Restriction digestion of the subcloning plasmid pUCE-TcdA-C2 (synthesized by Gene Company, containing the sequence of the first optimized combination Fragment 2), gel recovery to obtain the sequence of Fragment 2;
[0112] 2. Acquisition of the prokaryotic vector pET32b(+): design PCR primers to amplify the partial sequence of pET32b(+), and introduce an XhoI restriction site at one end. The PCR product purification kit recovers the product after the PCR reaction, and then releases the vector containing the cohesive end by single enzyme digestion with XhoI. Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com