Construction and application of co-expressed uptake transporters and drug-metabolizing enzyme models
A technology of uptake transporters and co-expression, applied in the biological field, can solve the problems of lack of comprehensive understanding of the synergy between drug transporters and metabolic enzymes, and few in vitro models
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Construction of recombinant plasmids pcDNA3.1(+) / OAT1 and pcDNA3.1 / Hygro(+) / CYP1A2
[0030] 1.1 Construction of recombinant plasmid pcDNA3.1(+) / OAT1
[0031] (1) Design two PCR primers according to the DNA sequence of human OAT1. The upstream and downstream primers respectively design a restriction site, EcoR I and Nhe I (underlined)
[0032] Upstream primer: 5’-CTA GCTAGC GACATGGCCTTTAATGACCTCCTGCAG-3';
[0033] Downstream primer: 5’-CAG GAATTC TCAGAGTCCATTCTTCTCTTGTGCTGA-3'.
[0034] (2) PCR was carried out using the pCMV6-OAT1 plasmid as a template, and the reaction system and reaction conditions are shown in Table 1.
[0035]
[0036]
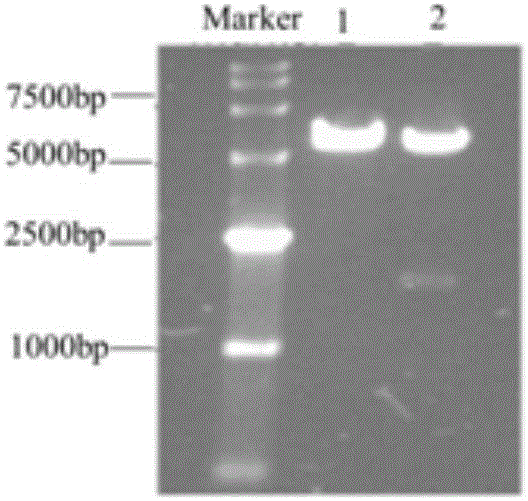
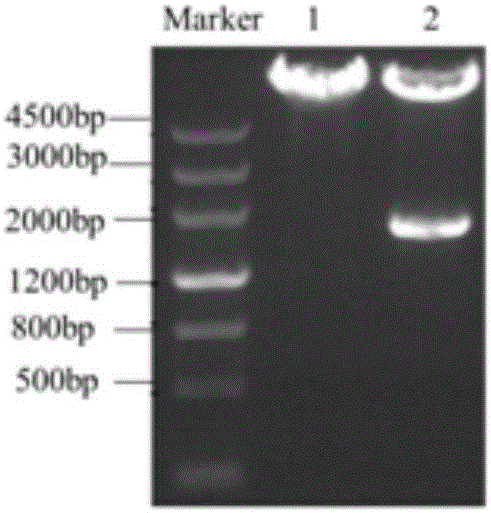
[0037] The PCR product was subjected to 1% agarose gel electrophoresis, and a 1.7kb electrophoresis band was obtained, which was consistent with the gene in the database OAT1 Same size.
[0038] (3) The target band was recovered by gel, and the pcDNA3.1 (+) plasmid and OAT1 gene fragment were respectively used E...
Embodiment 2
[0047] Example 2 MDCK cell transfection and OAT1 functional identification
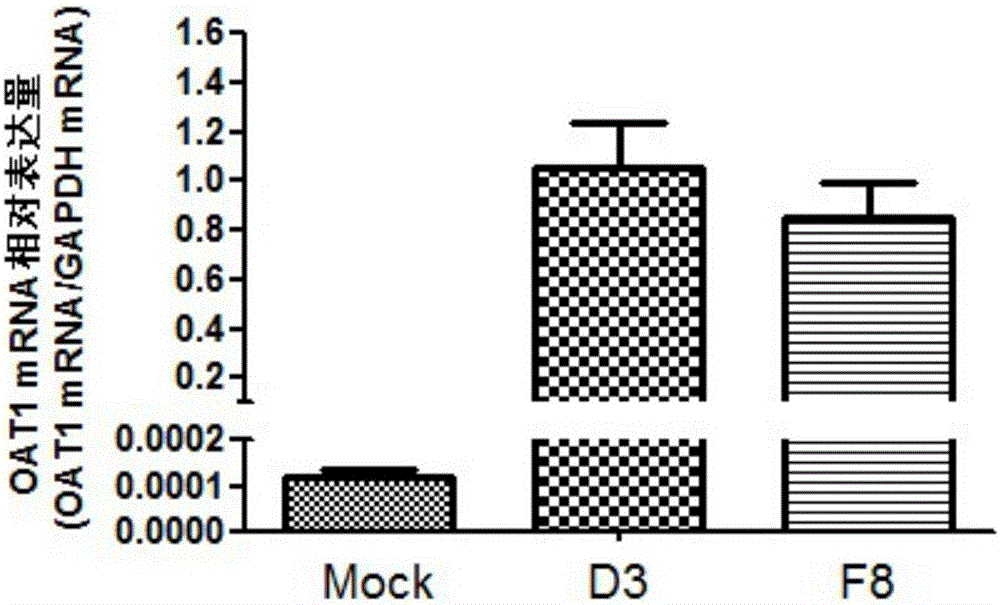
[0048] 2.1 Transfection of pcDNA3.1(+) / hOAT1 plasmid into MDCK cells
[0049] MDCK cells preserved in our laboratory were cultured in DMEM high-glucose medium containing 10% fetal bovine serum. Cells were placed at 37°C, 5% CO 2 cultured under conditions, passaged after trypsinization. The day before transfection, 2 × 10 per well 5 / mL for seeding and transfection when the cells reached 80% confluence. The pcDNA3.1(+) and pcDNA3.1(+) / hOAT1 plasmids used for transfection were extracted by a plasmid extraction kit and their concentrations were determined. Follow transfection reagent Lipofectamine TM 2000 MDCK cells were transfected according to the instructions. Add 100 μL of serum-free and antibiotic-free DMEM medium to two 1.5 mL Ependorff tubes. Add 6 μL of transfection reagent to one tube, and add 2-3 μg of plasmid to the other tube. The above two solutions were mixed and allowed to stand f...
Embodiment 3
[0065] Application of Example 3 in hOAT1 Inhibitor and Substrate Screening
[0066] Investigate glycyrrhizic acid, glycyrrhetinic acid, ligustrazine, ferulic acid, tanshinone, salvianolic acid A, salvianolic acid sodium, tanshinone I, salvianolic acid B, cryptotanshinone I, dihydrotanshinone I, aristolochic acid I, etc. Inhibitory effects of traditional Chinese medicine monomers on the uptake of fluorescent substrate 6-CFL by MDCK-OAT1 cells. The MDCK-OAT1 monoclonal cells were planted in 48-well plate, 1×105cell / well, and the accumulation experiment was carried out after 48h. The cells were divided into the control group and the test group, washed twice with HBSS solution, the cells in the control group were added with 200 μL HBSS solution containing 4 μmol / L 6-CFL, and the cells in the test group were added with 4 μmol / L 6-CFL and 100 μmol / L test drug (Probenecid was used as the inhibitor positive control with a concentration of 1 mmol / L) in 200 μL of HBSS solution, incubat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com