Method for randomly knocking out streptomyces genome DNA large fragment in vivo
A technology of large fragments and genomes, applied in the field of genetic engineering, can solve problems such as knockouts that cannot be knocked out with large fragments of genomes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
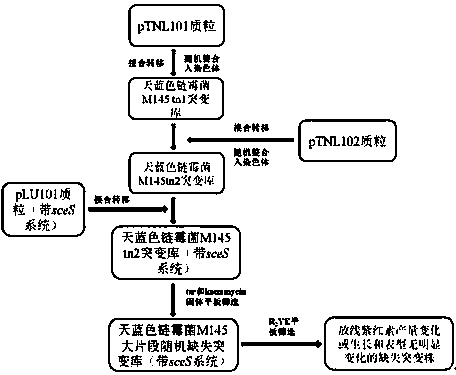
[0037] A method for randomly knocking out large fragments of Streptomyces genomic DNA in vivo, the flow chart is as follows figure 1 shown, including the following steps:
[0038] 1) Plasmid construction: Recombinant plasmids such as pTNL_101, pTNL_102, pTNL_103, pTNL_104 and pTNL_105 were completed on the basis of pDZY101 (CN102154268A) by PCR and conventional molecular cloning methods.
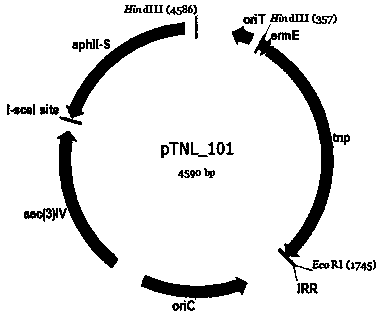
[0039] pTNL_101: Using pDZY101 as a template, PCR fragments with integrase, ermE promoter, IRR, oriT, apramycin resistance gene fragments and oriC were cloned into sceS The incomplete kanamycin resistance gene fragment at the restriction site (the name is aphII-S, the sequence is shown in SEQ ID NO.1), and its structure is shown in figure 2 shown.
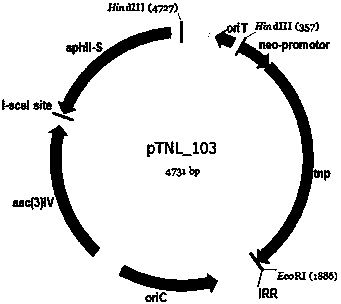
[0040] The construction methods of pTNL_103, pTNL_104 and pTNL_105 are the same as pTNL_101, the difference is that the promoters that start the transposase are different, which are neo promoter, hrdB Gene promoter and tcp830 inducible pro...
Embodiment 2
[0048] Transform the plasmids pTNL101 and pDZY101 into Escherichia coli ET12567, pick a single colony and insert them into 5mL LB liquid medium for overnight culture, transfer 1% of the inoculum to 100mL LB medium, and cultivate to OD at 37°C and 200rpm 600The value is about 0.3. After centrifugation, use 2×YT medium to wash twice, add 3mL 2×YT medium to mix the cells, and divide into 6 equal parts for later use. Use 6 mL of 2×YT medium, add 120 μL of Streptomyces coelicolor spore suspension, treat at 50°C for 10 minutes, divide into 6 equal parts, mix with the above bacteria, and then centrifuge to coat MS solid medium. After culturing at 30°C for 20 h, spread nalidixic acid and apramectin antibiotics on three pTNL101-transferred Streptomyces coelicolor M145 solid MS medium (final concentrations were 30 μg / mL and 50 μg / mL, respectively), 3 Nalidixic acid and apramectin (final concentrations of 30 μg / mL and 50 μg / mL, respectively) were coated on the solid MS medium of Streptom...
Embodiment 3
[0053] Transform the plasmids pTNL103 and pDZY101 into Escherichia coli ET12567, pick a single colony and insert them into 5mL LB liquid medium for overnight culture, transfer 1% of the inoculum to 100mL LB medium, and cultivate to OD at 37°C and 200rpm 600 The value is about 0.3. After centrifugation, use 2×YT medium to wash twice, add 3mL 2×YT medium to mix the cells, and divide into 6 equal parts for later use. Use 6 mL of 2×YT medium, add 120 μL of Streptomyces coelicolor spore suspension, treat at 50°C for 10 minutes, divide into 6 equal parts, mix with the above bacteria, and then centrifuge to coat MS solid medium. After culturing at 30°C for 20 h, spread nalidixic acid and apramectin antibiotics on three pTNL101-transferred Streptomyces coelicolor M145 solid MS medium (final concentrations were 30 μg / mL and 50 μg / mL, respectively), 3 Nalidixic acid and apramectin (final concentrations of 30 μg / mL and 50 μg / mL, respectively) were coated on the solid MS medium of Strepto...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com