Primers and probe for detecting corruption saccharomycete nucleotide fragments, detection method using the same and kit using the same
A detection method, yeast technology, applied in microorganism-based methods, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of difficulty in screening antibodies, inability to adapt, and high cost, to avoid false negative results, save money The effect of monitoring time and saving manpower and material resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] A method for detecting common spoilage yeast nucleotide fragments, comprising the steps of:
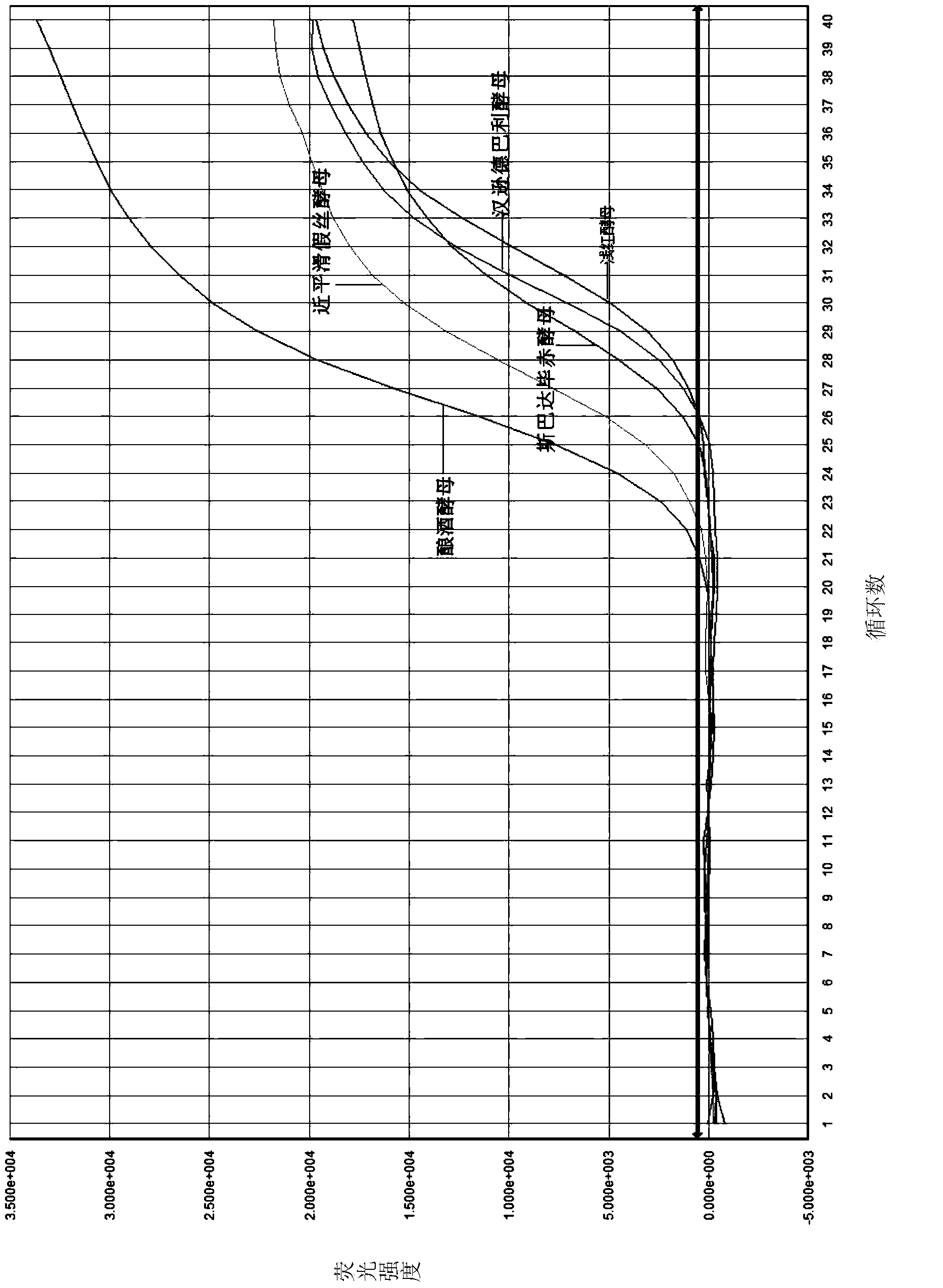
[0046] 1. Design of primers and probes: By comparing and analyzing the genome sequences of common spoilage yeasts, select a segment with no secondary structure and a high degree of conservation, and design multiple pairs of primers and probes. The length of the primers is generally about 20 bases , there is no complementary sequence between and within the primers. The optimal primer and probe sequence combinations are as follows:
[0047] Upstream primer Yeast F1:GAAGAGTCGAGTTGTTTGGGAA
[0048] Downstream primer Fungus R1: TCCTTCCCTTTCAACAATTTCAC
[0049] Probe Yeast Pb1: TGTACTTGTTCGCTATCGGTCTCTCGCC.
[0050]2. Establishment and optimization of the reaction system: The target region templates used in the establishment and optimization of the reaction system were obtained by the following method: Saccharomyces cerevisiae, Candida parapsilosis, Pichia sparta, Debari hansen Y...
Embodiment 2
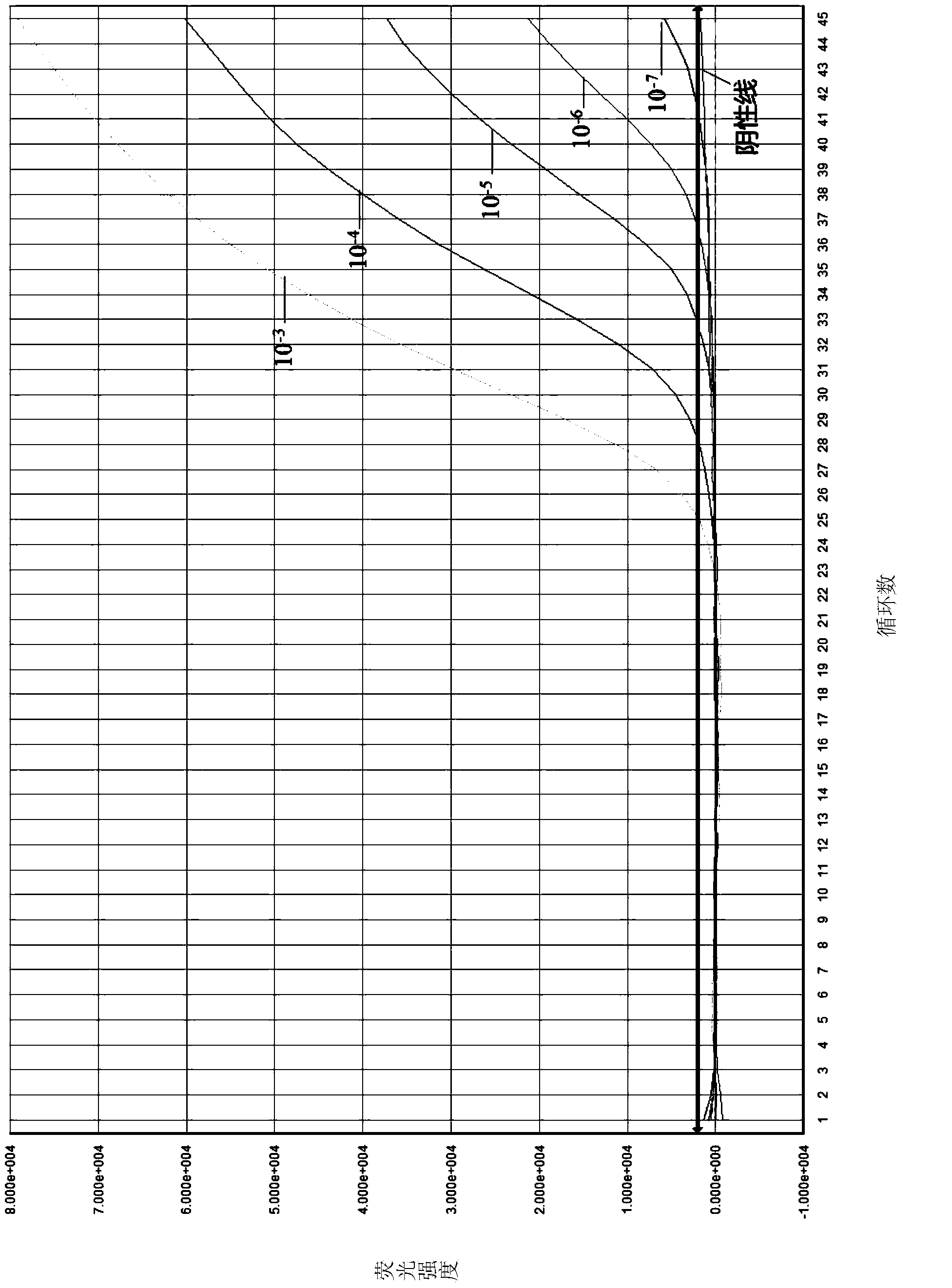
[0068] 1. Select the primer pair Yeast F1 / Fungus R1 primer and the probe Yeast Pb1, take the standard strain of Saccharomyces cerevisiae (CICC1001, purchased from the China Industrial Microbiology Culture Collection Management Center) and culture it in a selective medium for a certain period of time, and measure its OD value, estimate the number of bacteria, and then serially dilute 10 -1 , 10 -2 , 10 -3 , 10 -4 , 10 -5 , 10 -6 , 10 -7 , to study the sensitivity of the primer and probe system, take 10 -3 , 10 -4 , 10 -5 , 10 -6 , 10 -7 Plate counting, the amount of bacterial solution on the plate is 50ul / plate. and put 10 -3 , 10 -4 , 10 -5 , 10 -6 , 10 -7 Genomic DNA was extracted with the Takara Yeast Extraction Kit for the five gradient bacterial liquids. For specific steps, refer to the kit instruction manual.
[0069] 2. In the 18 μl detection kit, add 2 μl of each gradient yeast genomic DNA extracted above, and take 2 μl of DEPC water as a negative contro...
Embodiment 3
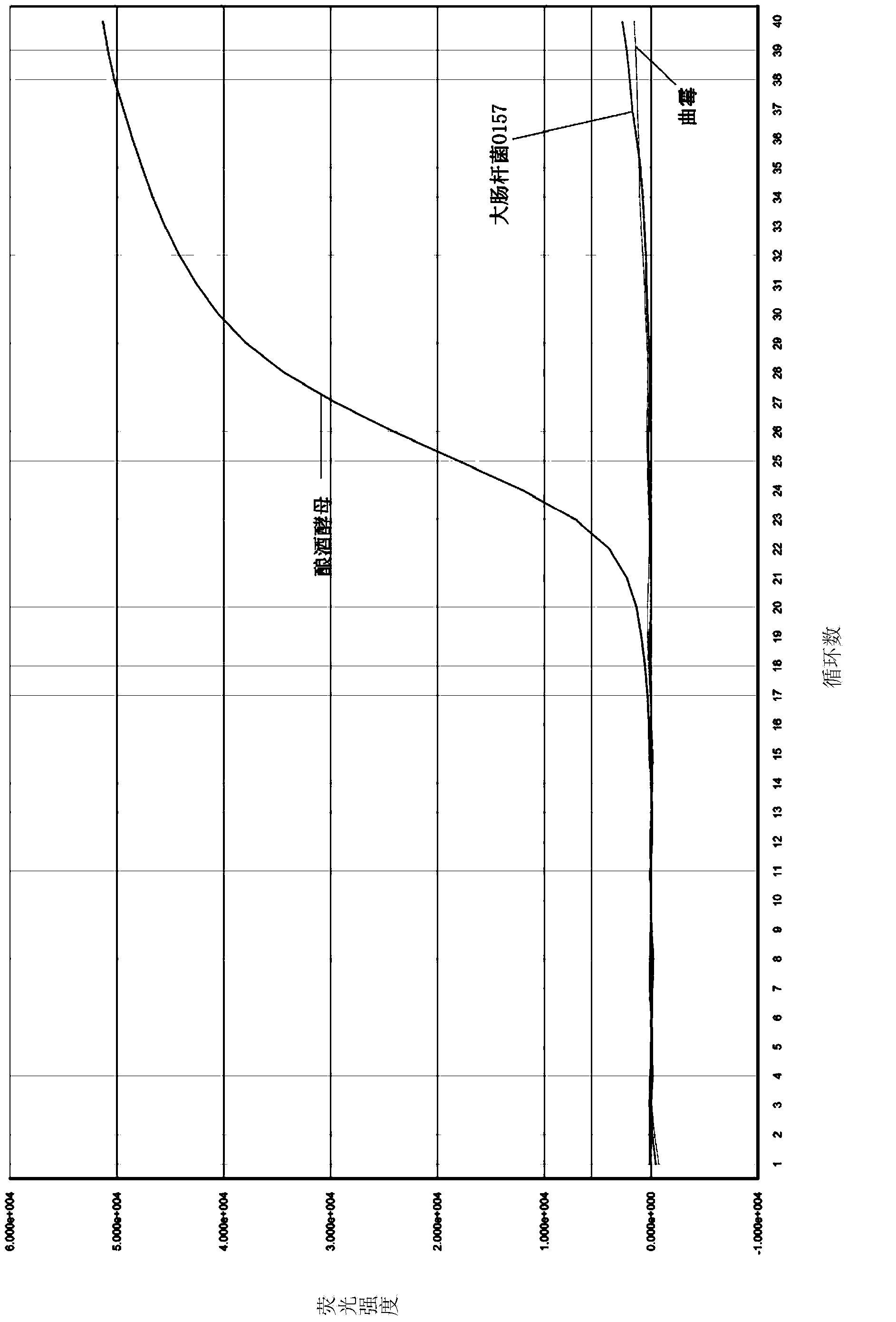
[0074] 1. Select primer pair Yeast F1 / Fungus R1 primer and probe Yeast Pb1, use bacteria Escherichia coli O157 (NCTC12900, provided by Zhuhai Entry-Exit Inspection and Quarantine Bureau) and mold Aspergillus (HG AO1, provided by Zhuhai Entry-Exit Inspection and Quarantine Bureau) The issue of specificity of this primer and probe system was investigated.
[0075] Genomic DNA was extracted from Escherichia coli O157 using the phenol-chloroform method, and the specific steps were as follows:
[0076] (1) Add the 9 pathogenic bacteria enrichment solutions (about 1 mL) to be tested into 1.5 mL centrifuge tubes, centrifuge at 12000 rpm for 5 minutes, and remove the supernatant;
[0077] (2) Add 700 μL of DNA Lysis Solution, mix well and resuspend, boil in water bath for 5 minutes;
[0078] (3) Add an equal volume of phenol-chloroform (V / V=1:1) solution, mix well and centrifuge at 13000rpm for 5 minutes;
[0079] (4) Transfer the supernatant into another 1.5mL centrifuge tube, add ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com