High-flux STR sequence core replication number detection method
A detection method and repeat number technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of limited number of detected loci, easy judgment, low throughput, etc., to reduce detection time and Cost, detection method is simple and fast, and the effect of small interference between reactions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Forensic identification STR detection based on biochip platform
[0041] This experiment is the application of the present invention in the high-throughput chip detection platform, which can realize the parallel detection of tens of thousands of detection sites. Compared with the current 96-channel parallel detection based on fluorescence capillary electrophoresis, the detection results obtained in a single run This also means that the detection time and detection cost of a single sample are greatly reduced.
[0042] The detection steps are:
[0043] (1) Preparation of on-chip detection sequence library:
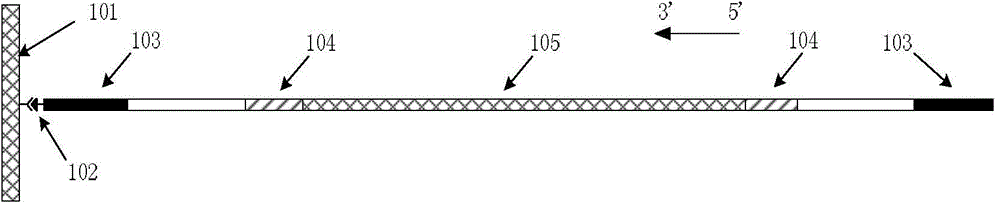
[0044] First, genomic DNA is extracted from the tissue to be tested. The 16 pairs of amplification primers shown in Table 1 were selected for multiple amplification of the corresponding loci in the genomic DNA. Among them, Amelogenin is the locus for detecting sex, and the other 15 are different STR loci.
[0045] The STR sequences to be detected in thi...
Embodiment 2
[0066] Example 2: Forensic identification based on high-throughput sequencing platform STR magnetic bead detection
[0067] This experiment is the application of the present invention in a high-throughput sequencing platform. Its detection throughput has been further improved, and parallel detection of millions of detection sites can be realized, which further reduces the detection time and detection time of a single sample. cost.
[0068] The detection steps are:
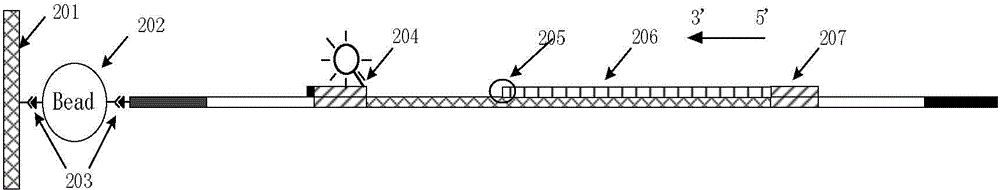
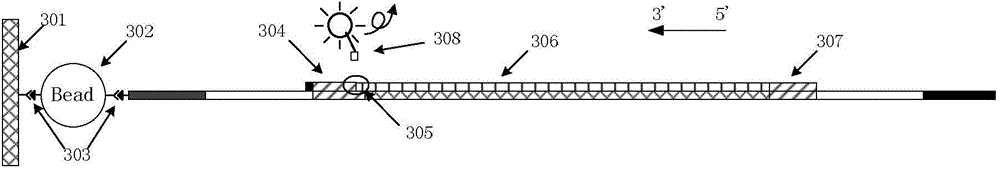
[0069] (1) Preparation of sequencing detection library:
[0070] Firstly, the genomic DNA was extracted from the tissue to be tested, and at the same time, magnetic beads connected with one of the 16 pairs of amplification primers shown in Table 1 were respectively prepared. The selection of unilateral primers was grouped according to the sequence characteristics of the core repeat region of the STR loci. Since the core repeating units of the 15 STR sequences in Table 1 all have a single base G (or C), they can ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com